当前位置:
X-MOL 学术
›
Environ. Sci. Technol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Integrating Genome-Resolved Metagenomics with Trait-Based Process Modeling to Determine Biokinetics of Distinct Nitrifying Communities within Activated Sludge
Environmental Science & Technology ( IF 10.8 ) Pub Date : 2022-08-05 , DOI: 10.1021/acs.est.2c02081 Pranav Sampara 1 , Yaqian Luo 1 , Xuan Lin 1 , Ryan M Ziels 1
Environmental Science & Technology ( IF 10.8 ) Pub Date : 2022-08-05 , DOI: 10.1021/acs.est.2c02081 Pranav Sampara 1 , Yaqian Luo 1 , Xuan Lin 1 , Ryan M Ziels 1
Affiliation
|
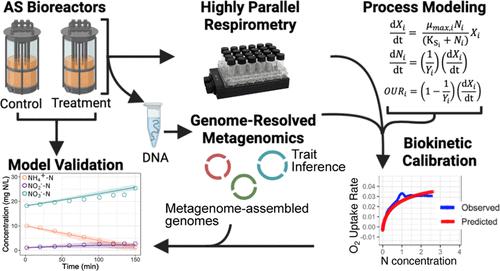
Conventional bioprocess models for wastewater treatment are based on aggregated bulk biomass concentrations and do not incorporate microbial physiological diversity. Such a broad aggregation of microbial functional groups can fail to predict ecosystem dynamics when high levels of physiological diversity exist within trophic guilds. For instance, functional diversity among nitrite-oxidizing bacteria (NOB) can obfuscate engineering strategies for their out-selection in activated sludge (AS), which is desirable to promote energy-efficient nitrogen removal. Here, we hypothesized that different NOB populations within AS can have different physiological traits that drive process performance, which we tested by estimating biokinetic growth parameters using a combination of highly replicated respirometry, genome-resolved metagenomics, and process modeling. A lab-scale AS reactor subjected to a selective pressure for over 90 days experienced resilience of NOB activity. We recovered three coexisting Nitrospira population genomes belonging to two sublineages, which exhibited distinct growth strategies and underwent a compositional shift following the selective pressure. A trait-based process model calibrated at the NOB genus level better predicted nitrite accumulation than a conventional process model calibrated at the NOB guild level. This work demonstrates that trait-based modeling can be leveraged to improve our prediction, control, and design of functionally diverse microbiomes driving key environmental biotechnologies.
中文翻译:

将基因组解析的宏基因组学与基于性状的过程建模相结合,以确定活性污泥中不同硝化群落的生物动力学
废水处理的传统生物过程模型基于聚集的大量生物质浓度,并且不考虑微生物的生理多样性。当营养群内存在高水平的生理多样性时,如此广泛的微生物功能群聚集可能无法预测生态系统动态。例如,亚硝酸盐氧化细菌(NOB)的功能多样性可能会扰乱其在活性污泥(AS)中的淘汰工程策略,而这对于促进节能脱氮是有利的。在这里,我们假设 AS 内的不同 NOB 群体可能具有不同的生理特征来驱动过程性能,我们通过结合高度重复的呼吸测量、基因组解析宏基因组学和过程建模来估计生物动力学生长参数来测试这一特征。承受选择压力超过 90 天的实验室规模 AS 反应器经历了 NOB 活性的恢复。我们恢复了属于两个亚系的三个共存的硝化螺旋菌群体基因组,它们表现出不同的生长策略,并在选择压力后经历了组成变化。在 NOB 属水平上校准的基于性状的过程模型比在 NOB 行会水平上校准的传统过程模型能更好地预测亚硝酸盐的积累。这项工作表明,基于性状的建模可用于改善我们对驱动关键环境生物技术的功能多样化微生物组的预测、控制和设计。
更新日期:2022-08-05
中文翻译:

将基因组解析的宏基因组学与基于性状的过程建模相结合,以确定活性污泥中不同硝化群落的生物动力学
废水处理的传统生物过程模型基于聚集的大量生物质浓度,并且不考虑微生物的生理多样性。当营养群内存在高水平的生理多样性时,如此广泛的微生物功能群聚集可能无法预测生态系统动态。例如,亚硝酸盐氧化细菌(NOB)的功能多样性可能会扰乱其在活性污泥(AS)中的淘汰工程策略,而这对于促进节能脱氮是有利的。在这里,我们假设 AS 内的不同 NOB 群体可能具有不同的生理特征来驱动过程性能,我们通过结合高度重复的呼吸测量、基因组解析宏基因组学和过程建模来估计生物动力学生长参数来测试这一特征。承受选择压力超过 90 天的实验室规模 AS 反应器经历了 NOB 活性的恢复。我们恢复了属于两个亚系的三个共存的硝化螺旋菌群体基因组,它们表现出不同的生长策略,并在选择压力后经历了组成变化。在 NOB 属水平上校准的基于性状的过程模型比在 NOB 行会水平上校准的传统过程模型能更好地预测亚硝酸盐的积累。这项工作表明,基于性状的建模可用于改善我们对驱动关键环境生物技术的功能多样化微生物组的预测、控制和设计。











































 京公网安备 11010802027423号
京公网安备 11010802027423号