Journal of Advanced Research ( IF 11.4 ) Pub Date : 2022-08-04 , DOI: 10.1016/j.jare.2022.07.007 Zhenfei Sun 1 , Yunlong Wang 2 , Zhaojian Song 3 , Hui Zhang 4 , Yuanda Wang 1 , Kunpeng Liu 1 , Min Ma 1 , Pan Wang 1 , Yaping Fang 2 , Detian Cai 3 , Guoliang Li 2 , Yuda Fang 1
|
Introduction
Polyploidy is a major force in plant evolution and the domestication of cultivated crops.
Objectives
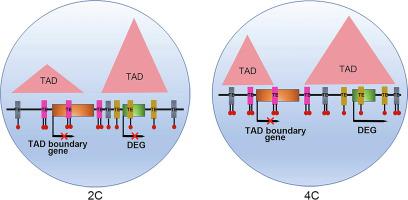
The study aimed to explore the relationship and underlying mechanism between three-dimensional (3D) chromatin organization and gene transcription upon rice genome duplication.
Methods
The 3D chromatin structures between diploid (2C) and autotetraploid (4C) rice were compared using high-throughput chromosome conformation capture (Hi-C) analysis. The study combined genetics, transcriptomics, whole-genome bisulfite sequencing (WGBS-seq) and 3D genomics approaches to uncover the mechanism for DNA methylation in modulating gene transcription through 3D chromatin architectures upon rice genome duplication.
Results
We found that 4C rice presents weakened intra-chromosomal interactions compared to its 2C progenitor in some chromosomes. In addition, we found that changes of 3D chromatin organizations including chromatin compartments, topologically associating domains (TADs), and loops, are uncorrelated with gene transcription. Moreover, DNA methylations in the regulatory sequences of genes in compartment A/B switched regions and TAD boundaries are unrelated to their expression. Importantly, although there was no significant difference in the methylation levels in transposable elements (TEs) in differentially expressed gene (DEG) and non-DEG promoters between 2C and 4C rice, we found that the hypermethylated TEs across genes in compartment A/B switched regions and TAD boundaries may suppress the expression of these genes.
Conclusion
The study proposed that the rice genome doubling might modulate TE methylation to buffer the effects of chromatin architecture on gene transcription in compartment A/B switched regions and TAD boundaries, resulting in the disconnection between 3D chromatin structure alteration and gene transcription upon rice genome duplication.
中文翻译:

转座因子中的 DNA 甲基化缓冲了水稻基因组复制时三维染色质组织与基因转录之间的联系
介绍
多倍体是植物进化和栽培作物驯化的主要力量。
目标
本研究旨在探索三维 (3D) 染色质组织与基因转录在水稻基因组复制过程中的关系和潜在机制。
方法
使用高通量染色体构象捕获 (Hi-C) 分析比较二倍体 (2C) 和同源四倍体 (4C) 水稻的 3D 染色质结构。该研究结合遗传学、转录组学、全基因组亚硫酸氢盐测序 (WGBS-seq) 和 3D 基因组学方法,揭示了水稻基因组复制时 DNA 甲基化通过 3D 染色质结构调节基因转录的机制。
结果
我们发现 4C 水稻在某些染色体中与其 2C 祖细胞相比表现出弱的染色体内相互作用。此外,我们发现 3D 染色质组织的变化,包括染色质区室、拓扑关联域 (TAD) 和环,与基因转录无关。此外,隔室 A/B 切换区域和 TAD 边界中基因调控序列中的 DNA 甲基化与它们的表达无关。重要的是,尽管 2C 和 4C 水稻之间差异表达基因 (DEG) 和非 DEG 启动子的转座因子 (TE) 的甲基化水平没有显着差异,但我们发现区室 A/B 中跨基因的高甲基化 TE 发生了切换区域和 TAD 边界可能会抑制这些基因的表达。
结论
该研究提出,水稻基因组加倍可能调节 TE 甲基化,以缓冲染色质结构对区室 A/B 转换区域和 TAD 边界中基因转录的影响,从而导致水稻基因组复制时 3D 染色质结构改变与基因转录之间的脱节。











































 京公网安备 11010802027423号
京公网安备 11010802027423号