当前位置:
X-MOL 学术
›
J. Am. Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
NEIL1 Recoding due to RNA Editing Impacts Lesion-Specific Recognition and Excision
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2022-08-02 , DOI: 10.1021/jacs.2c03625 Elizabeth R Lotsof 1 , Allison E Krajewski 2 , Brittany Anderson-Steele 1 , JohnPatrick Rogers 1 , Lanxin Zhang 2 , Jongchan Yeo 1 , Savannah G Conlon 1 , Amelia H Manlove 1 , Jeehiun K Lee 2 , Sheila S David 1
Journal of the American Chemical Society ( IF 14.4 ) Pub Date : 2022-08-02 , DOI: 10.1021/jacs.2c03625 Elizabeth R Lotsof 1 , Allison E Krajewski 2 , Brittany Anderson-Steele 1 , JohnPatrick Rogers 1 , Lanxin Zhang 2 , Jongchan Yeo 1 , Savannah G Conlon 1 , Amelia H Manlove 1 , Jeehiun K Lee 2 , Sheila S David 1
Affiliation
|
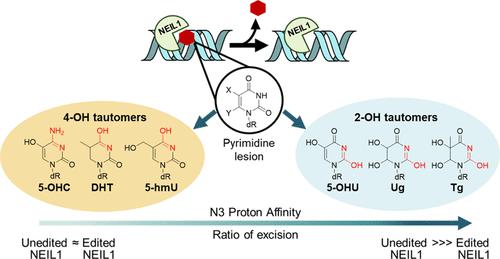
A-to-I RNA editing is widespread in human cells but is uncommon in the coding regions of proteins outside the nervous system. An unusual target for recoding by the adenosine deaminase ADAR1 is the pre-mRNA of the base excision DNA repair enzyme NEIL1 that results in the conversion of a lysine (K) to arginine (R) within the lesion recognition loop and alters substrate specificity. Differences in base removal by unedited (UE, K242) vs edited (Ed, R242) NEIL1 were evaluated using a series of oxidatively modified DNA bases to provide insight into the chemical and structural features of the lesion base that impact isoform-specific repair. We find that UE NEIL1 exhibits higher activity than Ed NEIL1 toward the removal of oxidized pyrimidines, such as thymine glycol, uracil glycol, 5-hydroxyuracil, and 5-hydroxymethyluracil. Gas-phase calculations indicate that the relative rates in excision track with the more stable lactim tautomer and the proton affinity of N3 of the base lesion. These trends support the contribution of tautomerization and N3 protonation in NEIL1 excision catalysis of these pyrimidine base lesions. Structurally similar but distinct substrate lesions, 5-hydroxycytosine and guanidinohydantoin, are more efficiently removed by the Ed NEIL1 isoform, consistent with the inherent differences in tautomerization, proton affinities, and lability. We also observed biphasic kinetic profiles and lack of complete base removal with specific combinations of the lesion and NEIL1 isoform, suggestive of multiple lesion binding modes. The complexity of NEIL1 isoform activity implies multiple roles for NEIL1 in safeguarding accurate repair and as an epigenetic regulator.
中文翻译:

由于 RNA 编辑影响病变特异性识别和切除而导致的 NEIL1 重新编码
A-to-I RNA 编辑在人类细胞中很普遍,但在神经系统外的蛋白质编码区域并不常见。腺苷脱氨酶 ADAR1 重新编码的一个不寻常目标是碱基切除 DNA 修复酶 NEIL1 的前体 mRNA,它会导致损伤识别环内的赖氨酸 (K) 转化为精氨酸 (R) 并改变底物特异性。使用一系列氧化修饰的 DNA 碱基评估未编辑的(UE,K242)与编辑的(Ed,R242)NEIL1 碱基去除的差异,以深入了解影响亚型特异性修复的损伤碱基的化学和结构特征。我们发现 UE NEIL1 在去除氧化嘧啶方面表现出比 Ed NEIL1 更高的活性,例如胸腺嘧啶乙二醇、尿嘧啶乙二醇、5-羟基尿嘧啶和 5-羟甲基尿嘧啶。气相计算表明切除中的相对速率与更稳定的内酰胺互变异构体和基础损伤的 N3 的质子亲和力相关。这些趋势支持互变异构化和 N3 质子化在这些嘧啶碱基损伤的 NEIL1 切除催化中的贡献。结构相似但不同的底物损伤、5-羟基胞嘧啶和胍基乙内酰脲更有效地被 Ed NEIL1 亚型去除,这与互变异构化、质子亲和力和不稳定性的固有差异一致。我们还观察到双相动力学特征和缺乏完整的碱基去除与病变和 NEIL1 亚型的特定组合,暗示多种病变结合模式。
更新日期:2022-08-02
中文翻译:

由于 RNA 编辑影响病变特异性识别和切除而导致的 NEIL1 重新编码
A-to-I RNA 编辑在人类细胞中很普遍,但在神经系统外的蛋白质编码区域并不常见。腺苷脱氨酶 ADAR1 重新编码的一个不寻常目标是碱基切除 DNA 修复酶 NEIL1 的前体 mRNA,它会导致损伤识别环内的赖氨酸 (K) 转化为精氨酸 (R) 并改变底物特异性。使用一系列氧化修饰的 DNA 碱基评估未编辑的(UE,K242)与编辑的(Ed,R242)NEIL1 碱基去除的差异,以深入了解影响亚型特异性修复的损伤碱基的化学和结构特征。我们发现 UE NEIL1 在去除氧化嘧啶方面表现出比 Ed NEIL1 更高的活性,例如胸腺嘧啶乙二醇、尿嘧啶乙二醇、5-羟基尿嘧啶和 5-羟甲基尿嘧啶。气相计算表明切除中的相对速率与更稳定的内酰胺互变异构体和基础损伤的 N3 的质子亲和力相关。这些趋势支持互变异构化和 N3 质子化在这些嘧啶碱基损伤的 NEIL1 切除催化中的贡献。结构相似但不同的底物损伤、5-羟基胞嘧啶和胍基乙内酰脲更有效地被 Ed NEIL1 亚型去除,这与互变异构化、质子亲和力和不稳定性的固有差异一致。我们还观察到双相动力学特征和缺乏完整的碱基去除与病变和 NEIL1 亚型的特定组合,暗示多种病变结合模式。











































 京公网安备 11010802027423号
京公网安备 11010802027423号