当前位置:
X-MOL 学术
›
J. Am. Chem. Soc.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
Cucurbit[7]uril Macrocyclic Sensors for Optical Fingerprinting: Predicting Protein Structural Changes to Identifying Disease-Specific Amyloid Assemblies
Journal of the American Chemical Society ( IF 15.0 ) Pub Date : 2022-08-01 , DOI: 10.1021/jacs.2c05969 Nilanjana Das Saha 1, 2 , Soumen Pradhan 2 , Ranjan Sasmal 1 , Aritra Sarkar 1 , Christian M Berač 3, 4 , Jonas C Kölsch 3 , Meenakshi Pahwa 2 , Sushanta Show 1 , Yves Rozenholc 5 , Zeki Topçu 5 , Vivien Alessandrini 6, 7 , Jean Guibourdenche 6, 7 , Vassilis Tsatsaris 6, 7 , Nathalie Gagey-Eilstein 6 , Sarit S Agasti 1, 2
Journal of the American Chemical Society ( IF 15.0 ) Pub Date : 2022-08-01 , DOI: 10.1021/jacs.2c05969 Nilanjana Das Saha 1, 2 , Soumen Pradhan 2 , Ranjan Sasmal 1 , Aritra Sarkar 1 , Christian M Berač 3, 4 , Jonas C Kölsch 3 , Meenakshi Pahwa 2 , Sushanta Show 1 , Yves Rozenholc 5 , Zeki Topçu 5 , Vivien Alessandrini 6, 7 , Jean Guibourdenche 6, 7 , Vassilis Tsatsaris 6, 7 , Nathalie Gagey-Eilstein 6 , Sarit S Agasti 1, 2
Affiliation
|
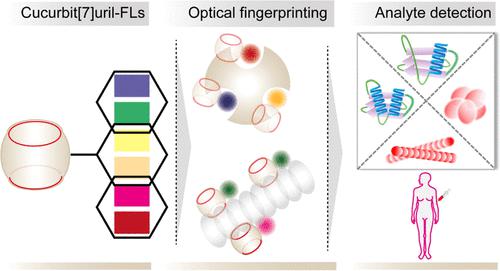
In a three-dimensional (3D) representation, each protein molecule displays a specific pattern of chemical and topological features, which are altered during its misfolding and aggregation pathway. Generating a recognizable fingerprint from such features could provide an enticing approach not only to identify these biomolecules but also to gain clues regarding their folding state and the occurrence of pathologically lethal misfolded aggregates. We report here a universal strategy to generate a fluorescent fingerprint from biomolecules by employing the pan-selective molecular recognition feature of a cucurbit[7]uril (CB[7]) macrocyclic receptor. We implemented a direct sensing strategy by covalently tethering CB[7] with a library of fluorescent reporters. When CB[7] recognizes the chemical and geometrical features of a biomolecule, it brings the tethered fluorophore into the vicinity, concomitantly reporting the nature of its binding microenvironment through a change in their optical signature. The photophysical properties of the fluorophores allow a multitude of probing modes, while their structural features provide additional binding diversity, generating a distinct fluorescence fingerprint from the biomolecule. We first used this strategy to rapidly discriminate a diverse range of protein analytes. The macrocyclic sensor was then applied to probe conformational changes in the protein structure and identify the formation of oligomeric and fibrillar species from misfolded proteins. Notably, the sensor system allowed us to differentiate between different self-assembled forms of the disease-specific amyloid-β (Aβ) aggregates and segregated them from other generic amyloid structures with a 100% identification accuracy. Ultimately, this sensor system predicted clinically relevant changes by fingerprinting serum samples from a cohort of pregnant women.
中文翻译:

用于光学指纹识别的 Cucurbit[7]uril 大环传感器:预测蛋白质结构变化以识别疾病特异性淀粉样蛋白组合物
在三维 (3D) 表示中,每个蛋白质分子都显示出特定的化学和拓扑特征模式,这些特征在其错误折叠和聚集途径中发生改变。从这些特征生成可识别的指纹可以提供一种诱人的方法,不仅可以识别这些生物分子,还可以获取有关它们的折叠状态和病理致死错误折叠聚集体发生的线索。我们在此报告了一种通用策略,通过使用葫芦[7] 脲 (CB[7]) 大环受体的泛选择性分子识别特征从生物分子中生成荧光指纹。我们通过将 CB[7] 与荧光报告基因库共价连接来实施直接传感策略。当 CB[7] 识别出生物分子的化学和几何特征时,它将系留的荧光团带到附近,同时通过改变它们的光学特征来报告其结合微环境的性质。荧光团的光物理特性允许多种探测模式,而它们的结构特征提供了额外的结合多样性,从生物分子中产生了独特的荧光指纹。我们首先使用这种策略来快速区分各种蛋白质分析物。然后应用大环传感器探测蛋白质结构的构象变化,并识别错误折叠蛋白质形成的寡聚和纤维状物质。尤其,传感器系统使我们能够区分疾病特异性淀粉样蛋白-β (Aβ) 聚集体的不同自组装形式,并将它们与其他通用淀粉样蛋白结构分离,并具有 100% 的识别准确度。最终,该传感器系统通过对一组孕妇的血清样本进行指纹识别来预测临床相关的变化。
更新日期:2022-08-01
中文翻译:

用于光学指纹识别的 Cucurbit[7]uril 大环传感器:预测蛋白质结构变化以识别疾病特异性淀粉样蛋白组合物
在三维 (3D) 表示中,每个蛋白质分子都显示出特定的化学和拓扑特征模式,这些特征在其错误折叠和聚集途径中发生改变。从这些特征生成可识别的指纹可以提供一种诱人的方法,不仅可以识别这些生物分子,还可以获取有关它们的折叠状态和病理致死错误折叠聚集体发生的线索。我们在此报告了一种通用策略,通过使用葫芦[7] 脲 (CB[7]) 大环受体的泛选择性分子识别特征从生物分子中生成荧光指纹。我们通过将 CB[7] 与荧光报告基因库共价连接来实施直接传感策略。当 CB[7] 识别出生物分子的化学和几何特征时,它将系留的荧光团带到附近,同时通过改变它们的光学特征来报告其结合微环境的性质。荧光团的光物理特性允许多种探测模式,而它们的结构特征提供了额外的结合多样性,从生物分子中产生了独特的荧光指纹。我们首先使用这种策略来快速区分各种蛋白质分析物。然后应用大环传感器探测蛋白质结构的构象变化,并识别错误折叠蛋白质形成的寡聚和纤维状物质。尤其,传感器系统使我们能够区分疾病特异性淀粉样蛋白-β (Aβ) 聚集体的不同自组装形式,并将它们与其他通用淀粉样蛋白结构分离,并具有 100% 的识别准确度。最终,该传感器系统通过对一组孕妇的血清样本进行指纹识别来预测临床相关的变化。



























 京公网安备 11010802027423号
京公网安备 11010802027423号