当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Addressing Missing Data in GC × GC Metabolomics: Identifying Missingness Type and Evaluating the Impact of Imputation Methods on Experimental Replication
Analytical Chemistry ( IF 6.7 ) Pub Date : 2022-07-26 , DOI: 10.1021/acs.analchem.1c04093 Trenton J Davis 1, 2 , Tarek R Firzli 3 , Emily A Higgins Keppler 1, 2 , Matthew Richardson 4, 5 , Heather D Bean 1, 2
Analytical Chemistry ( IF 6.7 ) Pub Date : 2022-07-26 , DOI: 10.1021/acs.analchem.1c04093 Trenton J Davis 1, 2 , Tarek R Firzli 3 , Emily A Higgins Keppler 1, 2 , Matthew Richardson 4, 5 , Heather D Bean 1, 2
Affiliation
|
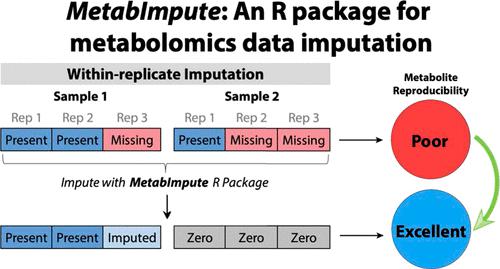
Missing data is a significant issue in metabolomics that is often neglected when conducting data preprocessing, particularly when it comes to imputation. This can have serious implications for downstream statistical analyses and lead to misleading or uninterpretable inferences. In this study, we aim to identify the primary types of missingness that affect untargeted metabolomics data and compare strategies for imputation using two real-world comprehensive two-dimensional gas chromatography (GC × GC) data sets. We also present these goals in the context of experimental replication whereby imputation is conducted in a within-replicate-based fashion─the first description and evaluation of this strategy─and introduce an R package MetabImpute to carry out these analyses. Our results conclude that, in these two GC × GC data sets, missingness was most likely of the missing at-random (MAR) and missing not-at-random (MNAR) types as opposed to missing completely at-random (MCAR). Gibbs sampler imputation and Random Forest gave the best results when imputing MAR and MNAR compared against single-value imputation (zero, minimum, mean, median, and half-minimum) and other more sophisticated approaches (Bayesian principal component analysis and quantile regression imputation for left-censored data). When samples are replicated, within-replicate imputation approaches led to an increase in the reproducibility of peak quantification compared to imputation that ignores replication, suggesting that imputing with respect to replication may preserve potentially important features in downstream analyses for biomarker discovery.
中文翻译:

解决 GC × GC 代谢组学中的缺失数据:识别缺失类型并评估插补方法对实验复制的影响
缺失数据是代谢组学中的一个重要问题,在进行数据预处理时经常被忽视,特别是在插补时。这可能会对下游统计分析产生严重影响,并导致误导性或无法解释的推论。在本研究中,我们的目标是确定影响非目标代谢组学数据的主要缺失类型,并使用两个真实世界的综合二维气相色谱 (GC × GC) 数据集比较插补策略。我们还在实验复制的背景下提出了这些目标,其中插补是以基于复制内的方式进行的(该策略的首次描述和评估),并引入 R 包 MetabImpute 来执行这些分析。我们的结果得出结论,在这两个 GC × GC 数据集中,缺失最有可能是随机缺失 (MAR) 和非随机缺失 (MNAR) 类型,而不是完全随机缺失 (MCAR)。与单值插补(零、最小值、平均值、中位数和半最小值)和其他更复杂的方法(贝叶斯主成分分析和分位数回归插补)相比,吉布斯采样器插补和随机森林在插补 MAR 和 MNAR 时给出了最佳结果左删失数据)。当样本被复制时,与忽略复制的插补相比,重复内插补方法导致峰定量的再现性提高,这表明与复制有关的插补可能会在生物标志物发现的下游分析中保留潜在的重要特征。
更新日期:2022-07-26
中文翻译:

解决 GC × GC 代谢组学中的缺失数据:识别缺失类型并评估插补方法对实验复制的影响
缺失数据是代谢组学中的一个重要问题,在进行数据预处理时经常被忽视,特别是在插补时。这可能会对下游统计分析产生严重影响,并导致误导性或无法解释的推论。在本研究中,我们的目标是确定影响非目标代谢组学数据的主要缺失类型,并使用两个真实世界的综合二维气相色谱 (GC × GC) 数据集比较插补策略。我们还在实验复制的背景下提出了这些目标,其中插补是以基于复制内的方式进行的(该策略的首次描述和评估),并引入 R 包 MetabImpute 来执行这些分析。我们的结果得出结论,在这两个 GC × GC 数据集中,缺失最有可能是随机缺失 (MAR) 和非随机缺失 (MNAR) 类型,而不是完全随机缺失 (MCAR)。与单值插补(零、最小值、平均值、中位数和半最小值)和其他更复杂的方法(贝叶斯主成分分析和分位数回归插补)相比,吉布斯采样器插补和随机森林在插补 MAR 和 MNAR 时给出了最佳结果左删失数据)。当样本被复制时,与忽略复制的插补相比,重复内插补方法导致峰定量的再现性提高,这表明与复制有关的插补可能会在生物标志物发现的下游分析中保留潜在的重要特征。











































 京公网安备 11010802027423号
京公网安备 11010802027423号