Journal of Advanced Research ( IF 11.4 ) Pub Date : 2022-07-25 , DOI: 10.1016/j.jare.2022.07.001 Miles McGibbon 1 , Sam Money-Kyrle 1 , Vincent Blay 2 , Douglas R Houston 1
|
Introduction
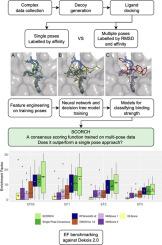
The discovery of a new drug is a costly and lengthy endeavour. The computational prediction of which small molecules can bind to a protein target can accelerate this process if the predictions are fast and accurate enough. Recent machine-learning scoring functions re-evaluate the output of molecular docking to achieve more accurate predictions. However, previous scoring functions were trained on crystalised protein-ligand complexes and datasets of decoys. The limited availability of crystal structures and biases in the decoy datasets can lower the performance of scoring functions.
Objectives
To address key limitations of previous scoring functions and thus improve the predictive performance of structure-based virtual screening.
Methods
A novel machine-learning scoring function was created, named SCORCH (Scoring COnsensus for RMSD-based Classification of Hits). To develop SCORCH, training data is augmented by considering multiple ligand poses and labelling poses based on their RMSD from the native pose. Decoy bias is addressed by generating property-matched decoys for each ligand and using the same methodology for preparing and docking decoys and ligands. A consensus of 3 different machine learning approaches is also used to improve performance.
Results
We find that multi-pose augmentation in SCORCH improves its docking power and screening power on independent benchmark datasets. SCORCH outperforms an equivalent scoring function trained on single poses, with a 1 % enrichment factor (EF) of 13.78 vs. 10.86 on 18 DEKOIS 2.0 targets and a mean native pose rank of 5.9 vs 30.4 on CSAR 2014. Additionally, SCORCH outperforms widely used scoring functions in virtual screening and pose prediction on independent benchmark datasets.
Conclusion
By rationally addressing key limitations of previous scoring functions, SCORCH improves the performance of virtual screening. SCORCH also provides an estimate of its uncertainty, which can help reduce the cost and time required for drug discovery.
中文翻译:

SCORCH:使用机器学习分类器、数据增强和不确定性估计改进基于结构的虚拟筛选
介绍
新药的发现是一项代价高昂且漫长的工作。如果预测足够快速和准确,则计算预测哪些小分子可以与蛋白质靶标结合可以加速这一过程。最近的机器学习评分函数重新评估分子对接的输出,以实现更准确的预测。然而,以前的评分函数是在结晶的蛋白质-配体复合物和诱饵数据集上训练的。晶体结构的有限可用性和诱饵数据集中的偏差会降低评分函数的性能。
目标
解决以前评分函数的关键局限性,从而提高基于结构的虚拟筛选的预测性能。
方法
创建了一个新的机器学习评分函数,名为 SCORCH(基于 RMSD 的命中分类的评分共识)。为了开发 SCORCH,通过考虑多个配体姿势和基于原始姿势的 RMSD 标记姿势来增强训练数据。通过为每个配体生成属性匹配的诱饵并使用相同的方法来准备和对接诱饵和配体来解决诱饵偏差。还使用 3 种不同机器学习方法的共识来提高性能。
结果
我们发现 SCORCH 中的多姿态增强提高了它在独立基准数据集上的对接能力和筛选能力。SCORCH 优于在单一姿势上训练的等效评分函数,在 18 个 DEKOIS 2.0 目标上的 1% 富集因子 (EF) 为 13.78 对比 10.86,在 CSAR 2014 上的平均原生姿势等级为 5.9 对比 30.4。此外,SCORCH 的表现优于广泛使用的虚拟筛选中的评分功能和独立基准数据集上的姿势预测。
结论
通过合理解决以前评分函数的关键局限性,SCORCH 提高了虚拟筛选的性能。SCORCH 还提供了对其不确定性的估计,这有助于降低药物发现所需的成本和时间。









































 京公网安备 11010802027423号
京公网安备 11010802027423号