Journal of Molecular Histology ( IF 2.9 ) Pub Date : 2022-07-23 , DOI: 10.1007/s10735-022-10082-w Ruizhen Sun 1 , Tiantian Gong 2 , Hui Liu 3 , Jingling Shen 1 , Bin Wu 1 , Qi Jiang 1 , Qi Wang 1 , Yue Zhang 1 , Lian Duan 1 , Jing Hu 1 , Qiuming Li 1 , Lei Lei 1 , Zhiyan Shan 1
|
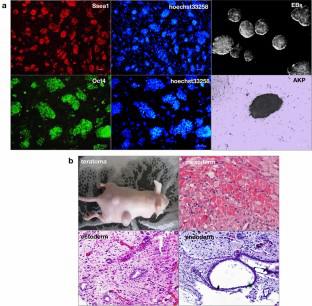
Differentiated cells can be reprogrammed to embryonic stem cell-like cells called induced pluripotent stem cells (iPSCs), in which the natural developmental differentiation process is reversed. It is unclear whether the multi-lineage cells can be isolated and identified during reprogramming. In the current study, we detected the expression of lineage markers, isolated neural lineages, and identified the related microRNAs during iPSC formation. Our results demonstrated that a neuroectoderm appeared earlier than mesoderm and definitive endoderm before forming colonies when mouse embryonic fibroblasts were subjected to iPSC formation using transcription factors (TFs). On day 3, the cells expressed Sox1 and Nestin and had ultrastructure consistent with the transition to identity neural germ layer lineage. Fluorescence-activated cell sorting analysis revealed a peak (40%) in neural progenitor marker–positive cells. When subsequently cultured in a neural precursor cell medium, these cells proliferated slowly, became round and aggregated, generating into neurons and glia. Genome-wide microRNA (miRNA) analysis identified 45 differentially regulated miRNAs. Molecular network analysis demonstrated that these miRNAs validated 6,047 experimental mRNA targets. The GO functional annotation analysis of mRNA targets revealed that most genes were related to neurogenesis, such as growth cone, neuronal cell body, neuron projection, and cell junction synapse. The network of protein–protein interactions was observed, which demonstrated that key nodes of neural lineage reprogramming-associated targets were Sall1, Foxa2, Nf2, Ctnnb1, Shh, and Bmpr1a. Therefore, these data suggested that TFs can drive the reprogramming of somatic cells towards a pluripotent state via neuroectoderm. Moreover, the neural lineage reprogramming system can address how miRNAs influence their target sites.
中文翻译:

在重编程过程中鉴定与神经胚层谱系特异性祖细胞相关的 microRNA
分化的细胞可以重新编程为胚胎干细胞样细胞,称为诱导多能干细胞 (iPSC),其中自然发育分化过程被逆转。目前尚不清楚在重编程期间是否可以分离和识别多谱系细胞。在目前的研究中,我们检测了谱系标记的表达,分离了神经谱系,并在 iPSC 形成过程中鉴定了相关的 microRNA。我们的研究结果表明,当使用转录因子 (TF) 对小鼠胚胎成纤维细胞进行 iPSC 形成时,神经外胚层在形成菌落之前出现早于中胚层和定形内胚层。第 3 天,细胞表达Sox1和Nestin并具有与向同一神经胚层谱系过渡一致的超微结构。荧光激活细胞分选分析显示神经祖细胞标记阳性细胞中有一个峰值 (40%)。当随后在神经前体细胞培养基中培养时,这些细胞增殖缓慢,变圆并聚集,生成神经元和神经胶质。全基因组 microRNA (miRNA) 分析确定了 45 种差异调节的 miRNA。分子网络分析表明,这些 miRNA 验证了 6,047 个实验性 mRNA 靶标。mRNA靶点的GO功能注释分析显示,大多数基因与神经发生有关,如生长锥、神经元胞体、神经元投射和细胞连接突触。观察到蛋白质-蛋白质相互作用的网络,Sall1、Foxa2、Nf2、Ctnnb1、Shh和Bmpr1a。因此,这些数据表明 TF 可以通过神经外胚层驱动体细胞重编程向多能状态。此外,神经谱系重编程系统可以解决 miRNA 如何影响其目标位点。











































 京公网安备 11010802027423号
京公网安备 11010802027423号