Medical Image Analysis ( IF 10.7 ) Pub Date : 2022-07-13 , DOI: 10.1016/j.media.2022.102536 Mina Jafari 1 , Susan Francis 2 , Jonathan M Garibaldi 1 , Xin Chen 1
|
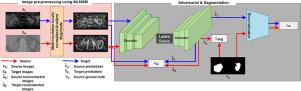
In medical image segmentation, supervised machine learning models trained using one image modality (e.g. computed tomography (CT)) are often prone to failure when applied to another image modality (e.g. magnetic resonance imaging (MRI)) even for the same organ. This is due to the significant intensity variations of different image modalities. In this paper, we propose a novel end-to-end deep neural network to achieve multi-modality image segmentation, where image labels of only one modality (source domain) are available for model training and the image labels for the other modality (target domain) are not available. In our method, a multi-resolution locally normalized gradient magnitude approach is firstly applied to images of both domains for minimizing the intensity discrepancy. Subsequently, a dual task encoder-decoder network including image segmentation and reconstruction is utilized to effectively adapt a segmentation network to the unlabeled target domain. Additionally, a shape constraint is imposed by leveraging adversarial learning. Finally, images from the target domain are segmented, as the network learns a consistent latent feature representation with shape awareness from both domains. We implement both 2D and 3D versions of our method, in which we evaluate CT and MRI images for kidney and cardiac tissue segmentation. For kidney, a public CT dataset (KiTS19, MICCAI 2019) and a local MRI dataset were utilized. The cardiac dataset was from the Multi-Modality Whole Heart Segmentation (MMWHS) challenge 2017. Experimental results reveal that our proposed method achieves significantly higher performance with a much lower model complexity in comparison with other state-of-the-art methods. More importantly, our method is also capable of producing superior segmentation results than other methods for images of an unseen target domain without model retraining. The code is available at GitHub (https://github.com/MinaJf/LMISA) to encourage method comparison and further research.
中文翻译:

LMISA:通过使用梯度幅度和形状约束进行域自适应的轻量级多模态图像分割网络
在医学图像分割中,使用一种图像模态(例如计算机断层扫描(CT))训练的监督机器学习模型在应用于另一种图像模态(例如磁共振成像(MRI))时通常容易失败,即使对于同一器官也是如此。这是由于不同图像模式的显着强度变化造成的。在本文中,我们提出了一种新颖的端到端深度神经网络来实现多模态图像分割,其中仅一种模态(源域)的图像标签可用于模型训练,而另一种模态(目标域)的图像标签可用于模型训练。域)不可用。在我们的方法中,首先将多分辨率局部归一化梯度幅度方法应用于两个域的图像,以最小化强度差异。随后,利用包括图像分割和重建的双任务编码器-解码器网络来有效地使分割网络适应未标记的目标域。此外,通过利用对抗性学习施加形状约束。最后,当网络从两个域中学习具有形状感知的一致的潜在特征表示时,来自目标域的图像被分割。我们实现了该方法的 2D 和 3D 版本,其中我们评估用于肾脏和心脏组织分割的 CT 和 MRI 图像。对于肾脏,使用了公共 CT 数据集(KiTS19、MICCAI 2019)和本地 MRI 数据集。心脏数据集来自 2017 年多模态全心脏分割 (MMWHS) 挑战赛。实验结果表明,与其他最先进的方法相比,我们提出的方法以更低的模型复杂度实现了显着更高的性能。 更重要的是,我们的方法还能够在无需模型重新训练的情况下,对不可见目标域的图像产生比其他方法更好的分割结果。该代码可在 GitHub (https://github.com/MinaJf/LMISA) 上获取,以鼓励方法比较和进一步研究。











































 京公网安备 11010802027423号
京公网安备 11010802027423号