当前位置:
X-MOL 学术
›
ACS Environ. Au
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A Combination of MALDI-TOF MS Proteomics and Species-Unique Biomarkers’ Discovery for Rapid Screening of Brucellosis
ACS Environmental Au ( IF 6.7 ) Pub Date : 2022-07-11 , DOI: 10.1021/jasms.2c00110 Hamideh Hamidi 1 , Ramin Bagheri Nejad 2 , Ali Es-haghi 2 , Alireza Ghassempour 1
ACS Environmental Au ( IF 6.7 ) Pub Date : 2022-07-11 , DOI: 10.1021/jasms.2c00110 Hamideh Hamidi 1 , Ramin Bagheri Nejad 2 , Ali Es-haghi 2 , Alireza Ghassempour 1
Affiliation
|
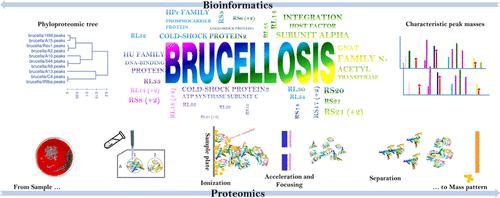
Brucellosis is considered to be a zoonotic infection with a predominant incidence in most parts of Iran that may even simply involve diagnostic laboratory personnel. In the present study, we apply matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF MS) for rapid and reliable discrimination of Brucella abortus and Brucella melitensis, based on proteomic mass patterns from chemically treated whole-cell analyses. Biomarkers of the low molecular weight proteome in the MALDI-TOF MS spectra were assigned to conserved ribosomal and structural protein families that were found in genome assemblies of B. abortus and B. melitensis in the NCBI database. Significant protein mass signals successfully mapped to ribosomal proteins and structural proteins, such as integration host factor subunit alpha, cold-shock proteins, HU family DNA-binding protein, ATP synthase subunit C, and GNAT family N-acetyltransferase, with specific biomarker peaks that have been identified for each virulent and vaccine strain. Web-accessible bioinformatics algorithms, with a robust data analysis workflow, followed by ribosomal and structural protein mapping, significantly enhanced the reliable assignment of key proteins and accurate identification of Brucella species. Furthermore, clinical samples were analyzed to confirm the most dominant protein biomarker candidates and their relevance for the identifications of B. melitensis and B. abortus. With proper optimization, we envision that the presented MALDI-TOF MS proteomics analyses, coupled with special usage of bioinformatics, could be used as a cost-efficient strategy for the diagnostics of brucellosis and introduce a reliable identification protocol for species of dangerous bacteria.
中文翻译:

MALDI-TOF MS 蛋白质组学和物种独特生物标志物的结合用于布鲁氏菌病的快速筛查
布鲁氏菌病被认为是一种人畜共患病,在伊朗大部分地区发病率很高,甚至可能只涉及诊断实验室人员。在本研究中,我们应用基质辅助激光解吸/电离飞行时间质谱 (MALDI-TOF MS) 快速可靠地区分流产布鲁氏菌和地中海布鲁氏菌,基于化学处理的全蛋白质组质量模式-细胞分析。MALDI-TOF MS 光谱中低分子量蛋白质组的生物标志物被分配到在流产芽孢杆菌和澳洲芽孢杆菌基因组组装中发现的保守核糖体和结构蛋白家族在 NCBI 数据库中。重要的蛋白质质量信号成功地映射到核糖体蛋白和结构蛋白,例如整合宿主因子亚基 α、冷休克蛋白、HU 家族 DNA 结合蛋白、ATP 合酶亚基 C 和 GNAT 家族 N-乙酰转移酶,具有特定的生物标志物峰已针对每种毒株和疫苗株进行了鉴定。可通过网络访问的生物信息学算法,以及强大的数据分析工作流程,以及随后的核糖体和结构蛋白作图,显着增强了关键蛋白的可靠分配和布鲁氏菌物种的准确鉴定。此外,对临床样本进行了分析,以确认最主要的蛋白质生物标志物候选物及其与B. melitensis和B.流产。通过适当的优化,我们设想所提出的 MALDI-TOF MS 蛋白质组学分析,再加上生物信息学的特殊用途,可用作布鲁氏菌病诊断的一种经济高效的策略,并为危险细菌的种类引入可靠的识别协议。
更新日期:2022-07-11
中文翻译:

MALDI-TOF MS 蛋白质组学和物种独特生物标志物的结合用于布鲁氏菌病的快速筛查
布鲁氏菌病被认为是一种人畜共患病,在伊朗大部分地区发病率很高,甚至可能只涉及诊断实验室人员。在本研究中,我们应用基质辅助激光解吸/电离飞行时间质谱 (MALDI-TOF MS) 快速可靠地区分流产布鲁氏菌和地中海布鲁氏菌,基于化学处理的全蛋白质组质量模式-细胞分析。MALDI-TOF MS 光谱中低分子量蛋白质组的生物标志物被分配到在流产芽孢杆菌和澳洲芽孢杆菌基因组组装中发现的保守核糖体和结构蛋白家族在 NCBI 数据库中。重要的蛋白质质量信号成功地映射到核糖体蛋白和结构蛋白,例如整合宿主因子亚基 α、冷休克蛋白、HU 家族 DNA 结合蛋白、ATP 合酶亚基 C 和 GNAT 家族 N-乙酰转移酶,具有特定的生物标志物峰已针对每种毒株和疫苗株进行了鉴定。可通过网络访问的生物信息学算法,以及强大的数据分析工作流程,以及随后的核糖体和结构蛋白作图,显着增强了关键蛋白的可靠分配和布鲁氏菌物种的准确鉴定。此外,对临床样本进行了分析,以确认最主要的蛋白质生物标志物候选物及其与B. melitensis和B.流产。通过适当的优化,我们设想所提出的 MALDI-TOF MS 蛋白质组学分析,再加上生物信息学的特殊用途,可用作布鲁氏菌病诊断的一种经济高效的策略,并为危险细菌的种类引入可靠的识别协议。











































 京公网安备 11010802027423号
京公网安备 11010802027423号