Structure ( IF 4.4 ) Pub Date : 2022-06-16 , DOI: 10.1016/j.str.2022.05.011 Chao Wang 1 , Frédéric Anglès 1 , William E Balch 1
|
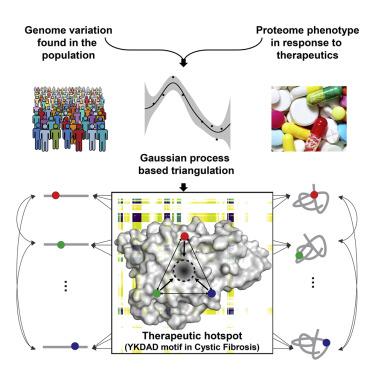
To understand mechanistically how the protein fold is shaped by therapeutics to inform precision management of disease, we developed variation-capture (VarC) mapping. VarC triangulates sparse sequence variation information found in the population using Gaussian process regression (GPR)-based machine learning to define the combined pairwise-residue interactions contributing to dynamic protein function in the individual in response to therapeutics. Using VarC mapping, we now reveal the pairwise-residue covariant relationships across the entire protein fold of cystic fibrosis (CF) transmembrane conductance regulator (CFTR) to define the molecular mechanisms of clinically approved CF chemical modulators. We discover an energetically destabilized covariant core containing a di-acidic YKDAD endoplasmic reticulum (ER) exit code that is only weakly corrected by current therapeutics. Our results illustrate that VarC provides a generalizable tool to triangulate information from genetic variation in the population to mechanistically discover therapeutic strategies that guide precision management of the individual.
中文翻译:

对人群中的变异进行三角测量以确定遗传病精准管理的机制
为了从机制上理解治疗方法如何塑造蛋白质折叠,从而为疾病的精确管理提供信息,我们开发了变异捕获(VarC)图谱。VarC 使用基于高斯过程回归 (GPR) 的机器学习对群体中发现的稀疏序列变异信息进行三角测量,以定义有助于个体响应治疗的动态蛋白质功能的组合成对残基相互作用。使用 VarC 图谱,我们现在揭示了囊性纤维化 (CF) 跨膜电导调节剂 (CFTR) 整个蛋白质折叠中的成对残基协变关系,以定义临床批准的 CF 化学调节剂的分子机制。我们发现了一个能量不稳定的协变核心,其中包含二酸性 YKDAD 内质网 (ER) 退出代码,目前的治疗只能微弱地纠正该代码。我们的结果表明,VarC 提供了一种通用工具,可以对群体遗传变异的信息进行三角测量,从而机械地发现指导个体精准管理的治疗策略。











































 京公网安备 11010802027423号
京公网安备 11010802027423号