Analytica Chimica Acta ( IF 5.7 ) Pub Date : 2022-06-15 , DOI: 10.1016/j.aca.2022.340094 Fatma Uysal Ciloglu 1 , Mehmet Hora 2 , Aycan Gundogdu 3 , Mehmet Kahraman 4 , Mahmut Tokmakci 5 , Omer Aydin 6
|
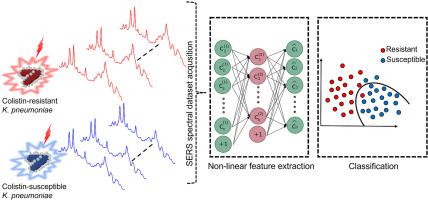
Colistin-resistant Klebsiella pneumoniae (ColR-Kp) causes high mortality rates since colistin is used as the last-line antibiotic against multi-drug resistant Gram-negative bacteria. To reduce infections and mortality rates caused by ColR-Kp fast and reliable detection techniques are vital. In this study, we used a label-free surface-enhanced Raman scattering (SERS)-based sensor with machine learning algorithms to discriminate colistin-resistant and susceptible strains of K. pneumoniae. A total of 16 K. pneumoniae strains were incubated in tryptic soy broth (TSB) for 4 h. Collected SERS spectra of ColR-Kp and colistin susceptible K. pneumoniae (ColS-Kp) have shown some spectral differences that hard to discriminate by the naked eye. To extract discriminative features from the dataset, autoencoder and principal component analysis (PCA) that extract features in a non-linear and linear manner, respectively were performed. Extracted features were fed into the support vector machine (SVM) classifier to discriminate K. pneumoniae strains. Classifier performance was evaluated by using features extracted by each feature extraction techniques. Classification results of SVM classifier with extracted features by an autoencoder (autoencoder-SVM) has shown better performance than SVM classifier with extracted features by PCA (PCA-SVM). The accuracy, sensitivity, specificity, and area under curve (AUC) value of the autoencoder-SVM model were found as 94%, 94.2%, 93.8%, and 0.98, respectively. Furthermore, the autoencoder-SVM model has demonstrated statistically significantly better classifier performance than PCA-SVM in terms of accuracy and AUC values. These results illustrate that non-linear features can be more discriminative than linear ones to determine SERS spectral data of antibiotic-resistant and susceptible bacteria. Our methodological approach enables rapid and high accuracy detection of ColR-Kp and ColS-Kp, suggesting that this can be a promising tool to limit colistin resistance.
中文翻译:

基于 SERS 的传感器与基于机器学习的有效特征提取技术用于快速检测耐粘菌素的肺炎克雷伯菌
多粘菌素耐药性肺炎克雷伯菌(ColR-Kp) 导致高死亡率,因为多粘菌素被用作对抗多重耐药革兰氏阴性菌的最后一线抗生素。为了降低由 ColR-Kp 引起的感染和死亡率,快速可靠的检测技术至关重要。在这项研究中,我们使用基于无标记表面增强拉曼散射 (SERS) 的传感器和机器学习算法来区分耐粘菌素和易感肺炎克雷伯菌菌株。将总共 16株肺炎克雷伯菌菌株在胰蛋白酶大豆肉汤 (TSB) 中培养 4 小时。收集的 ColR-Kp 和多粘菌素敏感的肺炎克雷伯菌的 SERS 光谱(ColS-Kp) 显示出一些肉眼难以辨别的光谱差异。为了从数据集中提取判别特征,分别执行了以非线性和线性方式提取特征的自动编码器和主成分分析 (PCA)。将提取的特征输入支持向量机 (SVM) 分类器以区分肺炎克雷伯菌菌株。通过使用每种特征提取技术提取的特征来评估分类器性能。通过自动编码器提取特征的支持向量机分类器(autoencoder-SVM)的分类结果显示出比通过 PCA 提取特征的支持向量机分类器(PCA-SVM)更好的性能。自编码器-SVM 模型的准确度、灵敏度、特异性和曲线下面积 (AUC) 值分别为 94%、94.2%、93.8% 和 0.98。此外,在准确性和 AUC 值方面,自编码器-SVM 模型在统计上证明了比 PCA-SVM 更好的分类器性能。这些结果表明,非线性特征比线性特征更能区分抗生素抗性和易感细菌的 SERS 光谱数据。











































 京公网安备 11010802027423号
京公网安备 11010802027423号