当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Development of OPLS-AA/M Parameters for Simulations of G Protein-Coupled Receptors and Other Membrane Proteins
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2022-06-10 , DOI: 10.1021/acs.jctc.2c00015 Michael J Robertson 1 , Georgios Skiniotis 1, 2
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2022-06-10 , DOI: 10.1021/acs.jctc.2c00015 Michael J Robertson 1 , Georgios Skiniotis 1, 2
Affiliation
|
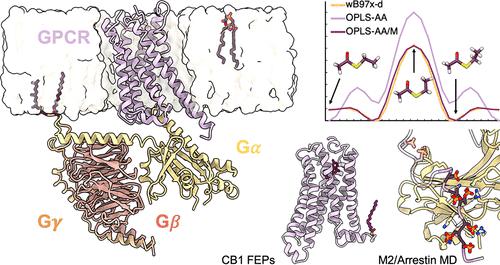
G protein-coupled receptors (GPCRs) and other membrane proteins are valuable drug targets, and their dynamic nature makes them attractive systems for study with molecular dynamics (MD) simulations and free energy approaches. Here, we report the development, implementation, and validation of OPLS-AA/M force field parameters to enable simulations of these systems. These efforts include the introduction of post-translational modifications including lipidations and phosphorylation. We also modify previously reported parameters for lipids to be more consistent with the OPLS-AA force field standard and extend their coverage. These new parameters are validated on a variety of test systems, with the results compared to high-level quantum mechanics calculations, experimental data, and simulations with other force fields. The results demonstrate that the new parameters reliably reproduce the behavior of membrane protein systems.
中文翻译:

开发用于模拟 G 蛋白偶联受体和其他膜蛋白的 OPLS-AA/M 参数
G 蛋白偶联受体 (GPCR) 和其他膜蛋白是有价值的药物靶点,它们的动态特性使其成为分子动力学 (MD) 模拟和自由能方法研究的有吸引力的系统。在这里,我们报告了 OPLS-AA/M 力场参数的开发、实施和验证,以实现对这些系统的模拟。这些努力包括引入翻译后修饰,包括脂化和磷酸化。我们还修改了先前报告的脂质参数,使其更符合 OPLS-AA 力场标准并扩大其覆盖范围。这些新参数在各种测试系统上进行了验证,并将结果与高级量子力学计算、实验数据和其他力场的模拟进行了比较。
更新日期:2022-06-10
中文翻译:

开发用于模拟 G 蛋白偶联受体和其他膜蛋白的 OPLS-AA/M 参数
G 蛋白偶联受体 (GPCR) 和其他膜蛋白是有价值的药物靶点,它们的动态特性使其成为分子动力学 (MD) 模拟和自由能方法研究的有吸引力的系统。在这里,我们报告了 OPLS-AA/M 力场参数的开发、实施和验证,以实现对这些系统的模拟。这些努力包括引入翻译后修饰,包括脂化和磷酸化。我们还修改了先前报告的脂质参数,使其更符合 OPLS-AA 力场标准并扩大其覆盖范围。这些新参数在各种测试系统上进行了验证,并将结果与高级量子力学计算、实验数据和其他力场的模拟进行了比较。









































 京公网安备 11010802027423号
京公网安备 11010802027423号