Medical Image Analysis ( IF 10.7 ) Pub Date : 2022-06-07 , DOI: 10.1016/j.media.2022.102512 Tim Ottens 1 , Sebastiano Barbieri 2 , Matthew R Orton 3 , Remy Klaassen 4 , Hanneke W M van Laarhoven 4 , Hans Crezee 5 , Aart J Nederveen 1 , Xiantong Zhen 6 , Oliver J Gurney-Champion 1
|
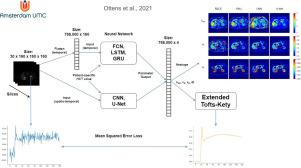
Dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) is an MRI technique for quantifying perfusion that can be used in clinical applications for classification of tumours and other types of diseases. Conventionally, the non-linear least squares (NLLS) methods is used for tracer-kinetic modelling of DCE data. However, despite promising results, NLLS suffers from long processing times (minutes-hours) and noisy parameter maps due to the non-convexity of the cost function. In this work, we investigated physics-informed deep neural networks for estimating physiological parameters from DCE-MRI signal-curves. Three voxel-wise temporal frameworks (FCN, LSTM, GRU) and two spatio-temporal frameworks (CNN, U-Net) were investigated. The accuracy and precision of parameter estimation by the temporal frameworks were evaluated in simulations. All networks showed higher precision than the NLLS. Specifically, the GRU showed to decrease the random error on by a factor of 4.8 with respect to the NLLS for noise (SD) of 1/20. The accuracy was better for the prediction of the parameter in all networks compared to the NLLS. The GRU and LSTM worked with arbitrary acquisition lengths. The GRU was selected for in vivo evaluation and compared to the spatio-temporal frameworks in 28 patients with pancreatic cancer. All neural network approaches showed less noisy parameter maps than the NLLS. The GRU had better test-retest repeatability than the NLLS for all three parameters and was able to detect one additional patient with significant changes in DCE parameters post chemo-radiotherapy. Although the U-Net and CNN had even better test-retest characteristics than the GRU, and were able to detect even more responders, they also showed potential systematic errors in the parameter maps. Therefore, we advise using our GRU framework for analysing DCE data.
中文翻译:

深度学习 DCE-MRI 参数估计:在胰腺癌中的应用
动态对比增强磁共振成像 (DCE-MRI) 是一种用于量化灌注的 MRI 技术,可用于临床应用以对肿瘤和其他类型的疾病进行分类。传统上,非线性最小二乘法 (NLLS) 方法用于 DCE 数据的示踪动力学建模。然而,尽管取得了可喜的结果,但由于成本函数的非凸性,NLLS 仍存在处理时间长(分钟-小时)和噪声参数图的问题。在这项工作中,我们研究了基于物理信息的深度神经网络,用于从 DCE-MRI 信号曲线估计生理参数。研究了三个体素时间框架(FCN、LSTM、GRU)和两个时空框架(CNN、U-Net)。在模拟中评估了时间框架的参数估计的准确性和精度。所有网络都显示出比 NLLS 更高的精度。具体来说,GRU 显示降低了随机误差对于 1/20 噪声 (SD) 的 NLLS,乘以 4.8 倍。预测的准确性更好与 NLLS 相比,所有网络中的参数。GRU 和 LSTM 使用任意采集长度。选择 GRU 进行体内评估,并与 28 名胰腺癌患者的时空框架进行比较。所有的神经网络方法都显示出比 NLLS 更少噪声的参数图。对于所有三个参数,GRU 具有比 NLLS 更好的重测可重复性,并且能够检测出另外一名在放化疗后 DCE 参数发生显着变化的患者。尽管 U-Net 和 CNN 比 GRU 具有更好的重测特性,并且能够检测到更多的响应者,但它们在参数图中也显示出潜在的系统误差。因此,我们建议使用我们的 GRU 框架来分析 DCE 数据。











































 京公网安备 11010802027423号
京公网安备 11010802027423号