Medical Image Analysis ( IF 10.9 ) Pub Date : 2022-05-27 , DOI: 10.1016/j.media.2022.102488 Silvia Seidlitz 1 , Jan Sellner 1 , Jan Odenthal 2 , Berkin Özdemir 3 , Alexander Studier-Fischer 3 , Samuel Knödler 3 , Leonardo Ayala 4 , Tim J Adler 5 , Hannes G Kenngott 6 , Minu Tizabi 7 , Martin Wagner 8 , Felix Nickel 8 , Beat P Müller-Stich 3 , Lena Maier-Hein 9
|
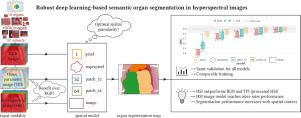
Semantic image segmentation is an important prerequisite for context-awareness and autonomous robotics in surgery. The state of the art has focused on conventional RGB video data acquired during minimally invasive surgery, but full-scene semantic segmentation based on spectral imaging data and obtained during open surgery has received almost no attention to date. To address this gap in the literature, we are investigating the following research questions based on hyperspectral imaging (HSI) data of pigs acquired in an open surgery setting: (1) What is an adequate representation of HSI data for neural network-based fully automated organ segmentation, especially with respect to the spatial granularity of the data (pixels vs. superpixels vs. patches vs. full images)? (2) Is there a benefit of using HSI data compared to other modalities, namely RGB data and processed HSI data (e.g. tissue parameters like oxygenation), when performing semantic organ segmentation? According to a comprehensive validation study based on 506 HSI images from 20 pigs, annotated with a total of 19 classes, deep learning-based segmentation performance increases — consistently across modalities — with the spatial context of the input data. Unprocessed HSI data offers an advantage over RGB data or processed data from the camera provider, with the advantage increasing with decreasing size of the input to the neural network. Maximum performance (HSI applied to whole images) yielded a mean DSC of 0.90 ((standard deviation (SD)) 0.04), which is in the range of the inter-rater variability (DSC of 0.89 ((standard deviation (SD)) 0.07)). We conclude that HSI could become a powerful image modality for fully-automatic surgical scene understanding with many advantages over traditional imaging, including the ability to recover additional functional tissue information. Our code and pre-trained models are available at https://github.com/IMSY-DKFZ/htc.
中文翻译:

高光谱图像中基于深度学习的鲁棒语义器官分割
语义图像分割是手术中上下文感知和自主机器人的重要先决条件。现有技术专注于在微创手术期间获取的传统 RGB 视频数据,但基于光谱成像数据和在开放手术期间获得的全场景语义分割迄今为止几乎没有受到关注。为了解决文献中的这一空白,我们正在研究基于在开放手术环境中获得的猪的高光谱成像 (HSI) 数据的以下研究问题:(1) 对于基于神经网络的全自动,HSI 数据的充分表示是什么器官分割,尤其是在数据的空间粒度方面(像素vs.超像素vs.补丁vs.全图)?(2) 在执行语义器官分割时,与其他模式(即 RGB 数据和处理后的 HSI 数据(例如氧合等组织参数)相比)使用 HSI 数据是否有好处?根据一项基于 20 头猪的 506 张 HSI 图像(共标注 19 个类别)的综合验证研究,基于深度学习的分割性能随着输入数据的空间上下文而不断提高——跨模式一致。未经处理的 HSI 数据比 RGB 数据或来自相机提供商的已处理数据具有优势,其优势随着神经网络输入大小的减小而增加。最大性能(HSI 应用于整个图像)产生的平均 DSC 为 0.90((标准偏差 (SD))0.04),这在评分者间变异性的范围内(DSC 为 0。89 ((标准偏差 (SD)) 0.07))。我们得出结论,HSI 可以成为全自动手术场景理解的强大图像模式,与传统成像相比具有许多优势,包括恢复额外功能组织信息的能力。我们的代码和预训练模型可在 https://github.com/IMSY-DKFZ/htc 获得。



























 京公网安备 11010802027423号
京公网安备 11010802027423号