当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
py-MCMD: Python Software for Performing Hybrid Monte Carlo/Molecular Dynamics Simulations with GOMC and NAMD
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2022-05-27 , DOI: 10.1021/acs.jctc.1c00911 Mohammad Soroush Barhaghi 1 , Brad Crawford 2 , Gregory Schwing 3 , David J Hardy 1 , John E Stone 1 , Loren Schwiebert 3 , Jeffrey Potoff 2 , Emad Tajkhorshid 1
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2022-05-27 , DOI: 10.1021/acs.jctc.1c00911 Mohammad Soroush Barhaghi 1 , Brad Crawford 2 , Gregory Schwing 3 , David J Hardy 1 , John E Stone 1 , Loren Schwiebert 3 , Jeffrey Potoff 2 , Emad Tajkhorshid 1
Affiliation
|
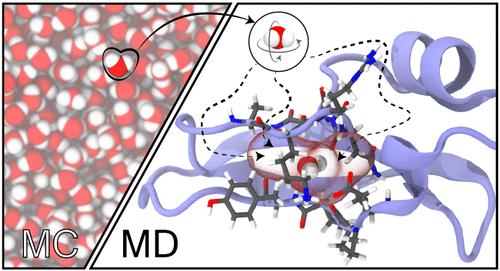
py-MCMD, an open-source Python software, provides a robust workflow layer that manages communication of relevant system information between the simulation engines NAMD and GOMC and generates coherent thermodynamic properties and trajectories for analysis. To validate the workflow and highlight its capabilities, hybrid Monte Carlo/molecular dynamics (MC/MD) simulations are performed for SPC/E water in the isobaric–isothermal (NPT) and grand canonical (GC) ensembles as well as with Gibbs ensemble Monte Carlo (GEMC). The hybrid MC/MD approach shows close agreement with reference MC simulations and has a computational efficiency that is 2 to 136 times greater than traditional Monte Carlo simulations. MC/MD simulations performed for water in a graphene slit pore illustrate significant gains in sampling efficiency when the coupled–decoupled configurational-bias MC (CD–CBMC) algorithm is used compared with simulations using a single unbiased random trial position. Simulations using CD–CBMC reach equilibrium with 25 times fewer cycles than simulations using a single unbiased random trial position, with a small increase in computational cost. In a more challenging application, hybrid grand canonical Monte Carlo/molecular dynamics (GCMC/MD) simulations are used to hydrate a buried binding pocket in bovine pancreatic trypsin inhibitor. Water occupancies produced by GCMC/MD simulations are in close agreement with crystallographically identified positions, and GCMC/MD simulations have a computational efficiency that is 5 times better than MD simulations. py-MCMD is available on GitHub at https://github.com/GOMC-WSU/py-MCMD.
中文翻译:

py-MCMD:使用 GOMC 和 NAMD 执行混合蒙特卡罗/分子动力学模拟的 Python 软件
py-MCMD 是一款开源 Python 软件,提供强大的工作流程层,用于管理模拟引擎 NAMD 和 GOMC 之间相关系统信息的通信,并生成连贯的热力学属性和轨迹以供分析。为了验证工作流程并突出其功能,对等压等温 (NPT) 和大正则 (GC) 系综以及吉布斯系综 Monte 中的 SPC/E 水进行了混合蒙特卡罗/分子动力学 (MC/MD )模拟卡罗(GEMC)。混合 MC/MD 方法与参考 MC 模拟非常一致,并且计算效率比传统蒙特卡罗模拟高 2 至 136 倍。对石墨烯狭缝孔中的水进行的 MC/MD 模拟表明,与使用单个无偏随机试验位置的模拟相比,使用耦合解耦配置偏差 MC (CD-CBMC) 算法时,采样效率显着提高。使用 CD-CBMC 的模拟达到平衡的周期数比使用单个无偏随机试验位置的模拟少 25 倍,计算成本略有增加。在更具挑战性的应用中,混合大正则蒙特卡罗/分子动力学 (GCMC/MD) 模拟用于水合牛胰腺胰蛋白酶抑制剂中的埋藏结合袋。GCMC/MD 模拟产生的水占有率与晶体学确定的位置非常一致,并且 GCMC/MD 模拟的计算效率比 MD 模拟高 5 倍。py-MCMD 可在 GitHub 上获取:https://github.com/GOMC-WSU/py-MCMD。
更新日期:2022-05-27
中文翻译:

py-MCMD:使用 GOMC 和 NAMD 执行混合蒙特卡罗/分子动力学模拟的 Python 软件
py-MCMD 是一款开源 Python 软件,提供强大的工作流程层,用于管理模拟引擎 NAMD 和 GOMC 之间相关系统信息的通信,并生成连贯的热力学属性和轨迹以供分析。为了验证工作流程并突出其功能,对等压等温 (NPT) 和大正则 (GC) 系综以及吉布斯系综 Monte 中的 SPC/E 水进行了混合蒙特卡罗/分子动力学 (MC/MD )模拟卡罗(GEMC)。混合 MC/MD 方法与参考 MC 模拟非常一致,并且计算效率比传统蒙特卡罗模拟高 2 至 136 倍。对石墨烯狭缝孔中的水进行的 MC/MD 模拟表明,与使用单个无偏随机试验位置的模拟相比,使用耦合解耦配置偏差 MC (CD-CBMC) 算法时,采样效率显着提高。使用 CD-CBMC 的模拟达到平衡的周期数比使用单个无偏随机试验位置的模拟少 25 倍,计算成本略有增加。在更具挑战性的应用中,混合大正则蒙特卡罗/分子动力学 (GCMC/MD) 模拟用于水合牛胰腺胰蛋白酶抑制剂中的埋藏结合袋。GCMC/MD 模拟产生的水占有率与晶体学确定的位置非常一致,并且 GCMC/MD 模拟的计算效率比 MD 模拟高 5 倍。py-MCMD 可在 GitHub 上获取:https://github.com/GOMC-WSU/py-MCMD。











































 京公网安备 11010802027423号
京公网安备 11010802027423号