当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
An In Silico Explainable Multiparameter Optimization Approach for De Novo Drug Design against Proteins from the Central Nervous System
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-05-17 , DOI: 10.1021/acs.jcim.2c00462 Navneet Bung 1 , Sowmya Ramaswamy Krishnan 1 , Arijit Roy 1
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2022-05-17 , DOI: 10.1021/acs.jcim.2c00462 Navneet Bung 1 , Sowmya Ramaswamy Krishnan 1 , Arijit Roy 1
Affiliation
|
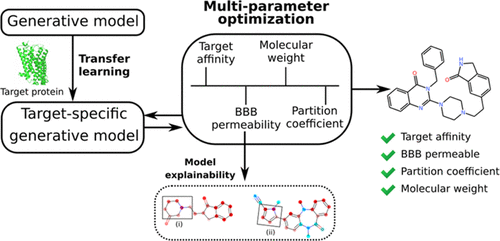
The aim of drug design and development is to produce a drug that can inhibit the target protein and possess a balanced physicochemical and toxicity profile. Traditionally, this is a multistep process where different parameters such as activity and physicochemical and pharmacokinetic properties are optimized sequentially, which often leads to high attrition rate during later stages of drug design and development. We have developed a deep learning-based de novo drug design method that can design novel small molecules by optimizing target specificity as well as multiple parameters (including late-stage parameters) in a single step. All possible combinations of parameters were optimized to understand the effect of each parameter over the other parameters. An explainable predictive model was used to identify the molecular fragments responsible for the property being optimized. The proposed method was applied against the human 5-hydroxy tryptamine receptor 1B (5-HT1B), a protein from the central nervous system (CNS). Various physicochemical properties specific to CNS drugs were considered along with the target specificity and blood–brain barrier permeability (BBBP), which act as an additional challenge for CNS drug delivery. The contribution of each parameter toward molecule design was identified by analyzing the properties of generated small molecules from optimization of all possible parameter combinations. The final optimized generative model was able to design similar inhibitors compared to known inhibitors of 5-HT1B. In addition, the functional groups of the generated small molecules that guide the BBBP predictive model were identified through feature attribution techniques.
中文翻译:

针对中枢神经系统蛋白质的从头药物设计的计算机可解释多参数优化方法
药物设计和开发的目的是生产一种能够抑制靶蛋白并具有平衡的物理化学和毒性特征的药物。传统上,这是一个多步骤的过程,其中不同的参数(如活性、物理化学和药代动力学特性)依次优化,这通常会导致药物设计和开发后期的高损耗率。我们开发了一种基于深度学习的de novo一种药物设计方法,可以通过一步优化目标特异性以及多个参数(包括后期参数)来设计新型小分子。所有可能的参数组合都经过优化,以了解每个参数对其他参数的影响。一个可解释的预测模型被用来识别负责被优化的特性的分子片段。所提出的方法适用于人类 5-羟基色胺受体 1B (5-HT1B),一种来自中枢神经系统 (CNS) 的蛋白质。考虑了 CNS 药物特有的各种物理化学特性以及靶标特异性和血脑屏障通透性 (BBBP),这是 CNS 药物递送的另一个挑战。通过对所有可能的参数组合的优化分析生成的小分子的性质,确定每个参数对分子设计的贡献。与已知的 5-HT1B 抑制剂相比,最终优化的生成模型能够设计出类似的抑制剂。此外,通过特征归因技术确定了指导 BBBP 预测模型的生成小分子的官能团。
更新日期:2022-05-17
中文翻译:

针对中枢神经系统蛋白质的从头药物设计的计算机可解释多参数优化方法
药物设计和开发的目的是生产一种能够抑制靶蛋白并具有平衡的物理化学和毒性特征的药物。传统上,这是一个多步骤的过程,其中不同的参数(如活性、物理化学和药代动力学特性)依次优化,这通常会导致药物设计和开发后期的高损耗率。我们开发了一种基于深度学习的de novo一种药物设计方法,可以通过一步优化目标特异性以及多个参数(包括后期参数)来设计新型小分子。所有可能的参数组合都经过优化,以了解每个参数对其他参数的影响。一个可解释的预测模型被用来识别负责被优化的特性的分子片段。所提出的方法适用于人类 5-羟基色胺受体 1B (5-HT1B),一种来自中枢神经系统 (CNS) 的蛋白质。考虑了 CNS 药物特有的各种物理化学特性以及靶标特异性和血脑屏障通透性 (BBBP),这是 CNS 药物递送的另一个挑战。通过对所有可能的参数组合的优化分析生成的小分子的性质,确定每个参数对分子设计的贡献。与已知的 5-HT1B 抑制剂相比,最终优化的生成模型能够设计出类似的抑制剂。此外,通过特征归因技术确定了指导 BBBP 预测模型的生成小分子的官能团。



























 京公网安备 11010802027423号
京公网安备 11010802027423号