当前位置:
X-MOL 学术
›
J. Comput. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Accelerating the 3D reference interaction site model theory of molecular solvation with treecode summation and cut-offs
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2022-05-14 , DOI: 10.1002/jcc.26889 Leighton Wilson 1 , Robert Krasny 1 , Tyler Luchko 2
Journal of Computational Chemistry ( IF 3.4 ) Pub Date : 2022-05-14 , DOI: 10.1002/jcc.26889 Leighton Wilson 1 , Robert Krasny 1 , Tyler Luchko 2
Affiliation
|
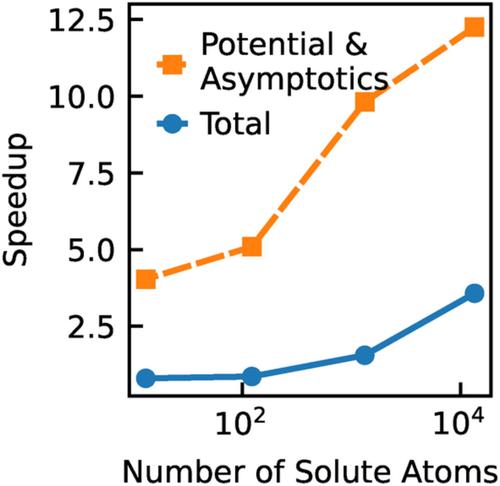
The 3D reference interaction site model (3D-RISM) of molecular solvation is a powerful tool for computing the equilibrium thermodynamics and density distributions of solvents, such as water and co-ions, around solute molecules. However, 3D-RISM solutions can be expensive to calculate, especially for proteins and other large molecules where calculating the potential energy between solute and solvent requires more than half the computation time. To address this problem, we have developed and implemented treecode summation for long-range interactions and analytically corrected cut-offs for short-range interactions to accelerate the potential energy and long-range asymptotics calculations in non-periodic 3D-RISM in the AmberTools molecular modeling suite. For the largest single protein considered in this work, tubulin, the total computation time was reduced by a factor of 4. In addition, parallel calculations with these new methods scale almost linearly and the iterative solver remains the largest impediment to parallel scaling. To demonstrate the utility of our approach for large systems, we used 3D-RISM to calculate the solvation thermodynamics and density distribution of 7-ring microtubule, consisting of 910 tubulin dimers, over 1.2 million atoms.
中文翻译:

使用树代码求和和截止加速分子溶剂化的 3D 参考相互作用位点模型理论
分子溶剂化的 3D 参考相互作用位点模型 (3D-RISM) 是计算溶质分子周围溶剂(例如水和共离子)的平衡热力学和密度分布的强大工具。然而,3D-RISM 解决方案的计算成本可能很高,尤其是对于蛋白质和其他大分子而言,计算溶质和溶剂之间的势能需要一半以上的计算时间。为了解决这个问题,我们开发并实现了长程相互作用的树代码求和和短程相互作用的解析校正截止,以加速 AmberTools 分子中非周期性 3D-RISM 中的势能和长程渐近计算建模套件。对于这项工作中考虑的最大的单一蛋白质,微管蛋白,总计算时间减少了 4 倍。此外,使用这些新方法的并行计算几乎可以线性扩展,而迭代求解器仍然是并行扩展的最大障碍。为了证明我们的方法在大型系统中的实用性,我们使用 3D-RISM 计算了 7 环微管的溶剂化热力学和密度分布,该微管由 910 个微管蛋白二聚体组成,超过 120 万个原子。
更新日期:2022-05-14
中文翻译:

使用树代码求和和截止加速分子溶剂化的 3D 参考相互作用位点模型理论
分子溶剂化的 3D 参考相互作用位点模型 (3D-RISM) 是计算溶质分子周围溶剂(例如水和共离子)的平衡热力学和密度分布的强大工具。然而,3D-RISM 解决方案的计算成本可能很高,尤其是对于蛋白质和其他大分子而言,计算溶质和溶剂之间的势能需要一半以上的计算时间。为了解决这个问题,我们开发并实现了长程相互作用的树代码求和和短程相互作用的解析校正截止,以加速 AmberTools 分子中非周期性 3D-RISM 中的势能和长程渐近计算建模套件。对于这项工作中考虑的最大的单一蛋白质,微管蛋白,总计算时间减少了 4 倍。此外,使用这些新方法的并行计算几乎可以线性扩展,而迭代求解器仍然是并行扩展的最大障碍。为了证明我们的方法在大型系统中的实用性,我们使用 3D-RISM 计算了 7 环微管的溶剂化热力学和密度分布,该微管由 910 个微管蛋白二聚体组成,超过 120 万个原子。











































 京公网安备 11010802027423号
京公网安备 11010802027423号