Structure ( IF 4.4 ) Pub Date : 2022-04-06 , DOI: 10.1016/j.str.2022.03.006 Helena Tossavainen 1 , Hasan Uğurlu 2 , Mikael Karjalainen 3 , Maarit Hellman 3 , Lina Antenucci 4 , Riku Fagerlund 2 , Kalle Saksela 2 , Perttu Permi 4
|
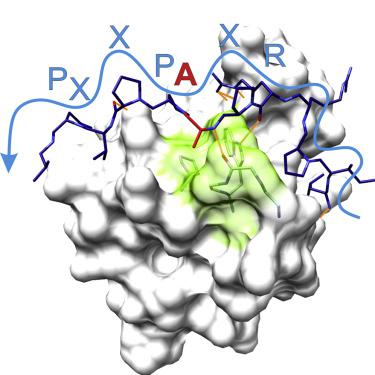
Class I SH3 domain-binding motifs generally comply with the consensus sequence [R/K]xØPxxP, the hydrophobic residue Ø being proline or leucine. We have studied the unusual Ø = Ala-specificity of SNX9 SH3 by determining its complex structure with a peptide present in eastern equine encephalitis virus (EEEV) nsP3. The structure revealed the length and composition of the n-Src loop as important factors determining specificity. We also compared the affinities of EEEV nsP3 peptide, its mutants, and cellular ligands to SNX9 SH3. These data suggest that nsP3 has evolved to minimize reduction of conformational entropy upon binding, hence acquiring stronger affinity, enabling takeover of SNX9. The RxAPxxP motif was also found in human T cell leukemia virus-1 (HTLV-1) Gag polyprotein. We found that this motif was required for efficient HTLV-1 infection, and that the specificity of SNX9 SH3 for the RxAPxxP core binding motif was importantly involved in this process.
中文翻译:

SNX9 SH3 与病毒配体复合的结构揭示了其对含丙氨酸的 I 类 SH3 基序的独特特异性的分子基础
I类SH3结构域结合基序通常符合共有序列[R/K]xØPxxP,疏水残基Ø是脯氨酸或亮氨酸。我们通过用东部马脑炎病毒 (EEEV) nsP3 中存在的肽确定其复杂结构,研究了 SNX9 SH3 的不寻常 Ø = Ala 特异性。该结构揭示了 n-Src 环的长度和组成是决定特异性的重要因素。我们还比较了 EEEV nsP3 肽、其突变体和细胞配体与 SNX9 SH3 的亲和力。这些数据表明,nsP3 已经进化到最大限度地减少结合后构象熵的减少,因此获得了更强的亲和力,从而能够接管 SNX9。RxAPxxP 基序也在人类 T 细胞白血病病毒 1 (HTLV-1) Gag 多蛋白中发现。我们发现这个基序是高效 HTLV-1 感染所必需的,











































 京公网安备 11010802027423号
京公网安备 11010802027423号