Journal of Advanced Research ( IF 11.4 ) Pub Date : 2022-03-31 , DOI: 10.1016/j.jare.2022.03.016 Duo Wang 1 , Yuanfeng Zhang 1 , Rui Li 1 , Jinming Li 1 , Rui Zhang 1
|
Introduction
Clinical precision oncology increasingly relies on accurate genome-wide profiling using large panel next generation sequencing; however, difficulties in accurate and consistent detection of somatic mutation from individual platforms and pipelines remain an open question.
Objectives
To obtain paired tumor–normal reference materials that can be effectively constructed and interchangeable with clinical samples, and evaluate the performance of 56 panels under routine testing conditions based on the reference samples.
Methods
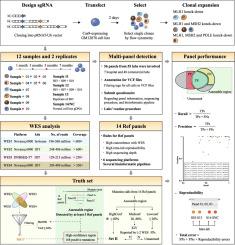
Genes involved in mismatch repair and DNA proofreading were knocked down using the CRISPR-Cas9 technology to accumulate somatic mutations in a defined GM12878 cell line. They were used as reference materials to comprehensively evaluate the reproducibility and accuracy of detection results of oncopanels and explore the potential influencing factors.
Results
In total, 14 paired tumor–normal reference DNA samples from engineered cell lines were prepared, and a reference dataset comprising 168 somatic mutations in a high-confidence region of 1.8 Mb were generated. For mutations with an allele frequency (AF) of more than 5% in reference samples, 56 panels collectively reported 1306 errors, including 729 false negatives (FNs), 179 false positives (FPs) and 398 reproducibility errors. The performance metric varied among panels with precision and recall ranging from 0.773 to 1 and 0.683 to 1, respectively. Incorrect and inadequate filtering accounted for a large proportion of false discovery (including FNs and FPs), while low-quality detection, cross-contamination and other sequencing errors during the wet bench process were other sources of FNs and FPs. In addition, low AF (<5%) considerably influenced the reproducibility and comparability among panels.
Conclusions
This study provided an integrated practice for developing reference standard to assess oncopanels in detecting somatic mutations and quantitatively revealed the source of detection errors. It will promote optimization, validation, and quality control among laboratories with potential applicability in clinical use.
中文翻译:

大面板下一代测序的一致性和可重复性:对具有错配修复和校对缺陷的参考材料进行体细胞突变检测的多实验室评估
介绍
临床精准肿瘤学越来越依赖于使用大面板下一代测序进行准确的全基因组分析;然而,从单个平台和管道准确和一致地检测体细胞突变的困难仍然是一个悬而未决的问题。
目标
获得可有效构建并可与临床样本互换的成对肿瘤-正常参考材料,并基于参考样本在常规测试条件下评估 56 组的性能。
方法
使用 CRISPR-Cas9 技术敲除错配修复和 DNA 校对中涉及的基因,从而在确定的 GM12878 细胞系中积累体细胞突变。将其作为参考资料,综合评价oncopanels检测结果的重现性和准确性,探索潜在的影响因素。
结果
总共制备了来自工程化细胞系的 14 对肿瘤-正常参考 DNA 样本,并生成了包含 1.8 Mb 高置信度区域中的 168 个体细胞突变的参考数据集。对于参考样本中等位基因频率 (AF) 超过 5% 的突变,56 个小组共报告了 1306 个错误,包括 729 个假阴性 (FN)、179 个假阳性 (FP) 和 398 个再现性错误。性能指标因面板而异,精度和召回率分别为 0.773 到 1 和 0.683 到 1。不正确和不充分的过滤是错误发现(包括 FN 和 FP)的很大一部分原因,而湿工作台过程中的低质量检测、交叉污染和其他测序错误是 FN 和 FP 的其他来源。此外,低 AF (<
结论
本研究为开发参考标准以评估 oncopanels 在检测体细胞突变方面提供了综合实践,并定量揭示了检测误差的来源。它将促进具有临床应用潜力的实验室之间的优化、验证和质量控制。











































 京公网安备 11010802027423号
京公网安备 11010802027423号