Journal of Environmental Chemical Engineering ( IF 7.4 ) Pub Date : 2022-01-19 , DOI: 10.1016/j.jece.2022.107219 Muhammad Ibrar 1, 2, 3, 4 , Xuewei Yang 1, 3
|
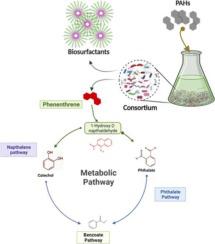
Due to substantial metabolic burden and low carbon fluxes, a single microorganism cannot utilize multiple classes of hydrocarbons typically found in the polluted environment. A microbial consortium comprising biosurfactants producing strains and hydrocarbon degradative strains could have promising bioremediation capabilities. To address this question, bacteria from non-contaminated soil were enriched in a medium containing polyaromatic hydrocarbons. The metabolic potential and pathways of PAHs degrading bacterial consortia (CON-PAH) that drive energy and carbon from PAHs assimilation were elucidated by metagenomic analysis. CON-PAH degrade more than 90% of PAHs (naphthalene, phenanthrene, and pyrene) with concomitant biosurfactants production. Tween-20 and glucose as a co-substrate suppress the PAHs degradation. Six high-quality metagenome-assembled genomes (MAGs) were assembled with an estimated size of 3.8–6.5 Mb from metagenomic data of CON-PAH. The draft genome of Deliftia tsuruhatensis and Pseudomonas putida contain the entire benzoate and near-complete naphthalene and phenanthrene degradation pathways. For the first time, we reconstruct PAH degrading pathway in D. tsuruhatensis sp. The metagenomic reconstruction of bins indicates the cross-feeding of bacteria within the consortium and suggests that complete PAH degradation pathways may not be obligatory for individual bacteria in the consortium. Biosurfactants structure elucidation via LC/MS showed lipopeptide containing the heptapeptides with different lipid moieties. This work will deepen our understanding of the metabolic capacity of microbial communities and provide new ideas for the engineering of artificial microbial consortia.
中文翻译:

重建富集菌群中的多环芳烃降解途径及其生物表面活性剂表征
由于大量的代谢负担和低碳通量,单一微生物不能利用通常在污染环境中发现的多种碳氢化合物。由产生生物表面活性剂的菌株和烃降解菌株组成的微生物联合体可能具有很有前景的生物修复能力。为了解决这个问题,来自未受污染土壤的细菌在含有多环芳烃的培养基中富集。通过宏基因组分析阐明了多环芳烃降解细菌聚生体 (CON-PAH) 的代谢潜力和途径,这些途径驱动了多环芳烃同化过程中的能量和碳。CON-PAH 可降解 90% 以上的 PAH(萘、菲和芘),同时产生生物表面活性剂。Tween-20 和葡萄糖作为共底物抑制 PAHs 降解。从 CON-PAH 的宏基因组数据中组装了六个高质量的宏基因组组装基因组 (MAG),估计大小为 3.8-6.5 Mb。基因组草图Deliftia tsuruhatensis和Pseudomonas putida包含完整的苯甲酸盐和近乎完整的萘和菲降解途径。我们首次在D. tsuruhatensis sp. 中重建 PAH 降解途径。垃圾箱的宏基因组重建表明菌群内细菌的交叉喂养,并表明完整的多环芳烃降解途径可能对菌群中的单个细菌不是强制性的。通过 LC/MS 进行的生物表面活性剂结构解析显示脂肽含有具有不同脂质部分的七肽。这项工作将加深我们对微生物群落代谢能力的理解,并为人工微生物群落的工程化提供新思路。











































 京公网安备 11010802027423号
京公网安备 11010802027423号