Cell ( IF 45.5 ) Pub Date : 2021-12-01 , DOI: 10.1016/j.cell.2021.11.012 Hyun Jung Oh 1 , Rodrigo Aguilar 1 , Barry Kesner 1 , Hun-Goo Lee 1 , Andrea J Kriz 1 , Hsueh-Ping Chu 1 , Jeannie T Lee 1
|
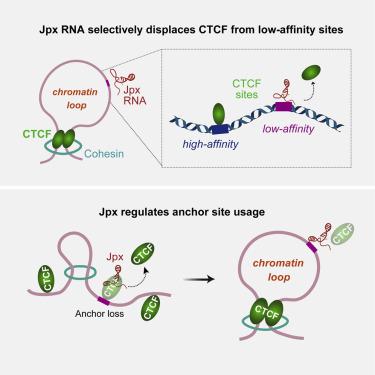
Chromosome loops shift dynamically during development, homeostasis, and disease. CCCTC-binding factor (CTCF) is known to anchor loops and construct 3D genomes, but how anchor sites are selected is not yet understood. Here, we unveil Jpx RNA as a determinant of anchor selectivity. Jpx RNA targets thousands of genomic sites, preferentially binding promoters of active genes. Depleting Jpx RNA causes ectopic CTCF binding, massive shifts in chromosome looping, and downregulation of >700 Jpx target genes. Without Jpx, thousands of lost loops are replaced by de novo loops anchored by ectopic CTCF sites. Although Jpx controls CTCF binding on a genome-wide basis, it acts selectively at the subset of developmentally sensitive CTCF sites. Specifically, Jpx targets low-affinity CTCF motifs and displaces CTCF protein through competitive inhibition. We conclude that Jpx acts as a CTCF release factor and shapes the 3D genome by regulating anchor site usage.
中文翻译:

Jpx RNA 调节 CTCF 锚定位点选择和染色体环的形成
染色体环在发育、稳态和疾病期间动态变化。已知 CCCTC 结合因子 (CTCF) 可锚定环并构建 3D 基因组,但尚不清楚如何选择锚定位点。在这里,我们揭示了 Jpx RNA 作为锚定选择性的决定因素。Jpx RNA 靶向数千个基因组位点,优先结合活性基因的启动子。耗尽 Jpx RNA 会导致异位 CTCF 结合、染色体环的大量移动以及 >700 Jpx 靶基因的下调。没有 Jpx,数千个丢失的循环被从头替换由异位 CTCF 站点锚定的循环。尽管 Jpx 在全基因组基础上控制 CTCF 结合,但它选择性地作用于发育敏感的 CTCF 位点子集。具体来说,Jpx 靶向低亲和力 CTCF 基序并通过竞争性抑制取代 CTCF 蛋白。我们得出结论,Jpx 作为 CTCF 释放因子并通过调节锚位点使用来塑造 3D 基因组。











































 京公网安备 11010802027423号
京公网安备 11010802027423号