Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Demonstration of elementary functions via DNA algorithmic self-assembly
Nanoscale ( IF 5.8 ) Pub Date : 2021-10-28 , DOI: 10.1039/d1nr05055a Muhammad Tayyab Raza 1 , Anshula Tandon 1 , Suyoun Park 1 , Sungjin Lee 1 , Thi Bich Ngoc Nguyen 1 , Thi Hong Nhung Vu 1 , Soojin Jo 1 , Yeonju Nam 1 , Sohee Jeon 2 , Jun-Ho Jeong 2, 3 , Sung Ha Park 1
Nanoscale ( IF 5.8 ) Pub Date : 2021-10-28 , DOI: 10.1039/d1nr05055a Muhammad Tayyab Raza 1 , Anshula Tandon 1 , Suyoun Park 1 , Sungjin Lee 1 , Thi Bich Ngoc Nguyen 1 , Thi Hong Nhung Vu 1 , Soojin Jo 1 , Yeonju Nam 1 , Sohee Jeon 2 , Jun-Ho Jeong 2, 3 , Sung Ha Park 1
Affiliation
|
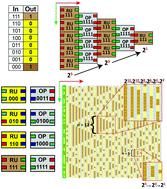
Target-oriented cellular automata with computation are the primary challenge in the field of DNA algorithmic self-assembly in connection with specific rules. We investigate the feasibility of using the principle of cellular automata for mathematical subjects by using specific logic gates that can be implemented into DNA building blocks. Here, we connect the following five representative elementary functions: (i) enumeration of multiples of 2, 3, and 4 (demonstrated via R094, R062, and R190 in 3-input/1-output logic rules); (ii) the remainder of 0 and 1 (R132); (iii) powers of 2 (R129); (iv) ceiling function for n/2 and n/4 (R152 and R144); and (v) analogous pattern of annihilation (R184) to DNA algorithmic patterns formed by specific rules. After designing the abstract building blocks and simulating the generation of algorithmic lattices, we conducted an experiment as follows: designing of DNA tiles with specific sticky ends, construction of DNA lattices via a two-step annealing method, and verification of expected algorithmic patterns on a given DNA lattice using an atomic force microscope (AFM). We observed representative patterns, such as horizontal and diagonal stripes and embedded triangles, on the given algorithmic lattices. The average error rates of individual rules are in the range of 8.8% (R184) to 11.9% (R062), and the average error rate for all the rules was 10.6%. Interpretation of elementary functions demonstrated through DNA algorithmic patterns could be extended to more complicated functions, which may lead to new insights for achieving the final answers of functions with experimentally obtained patterns.
中文翻译:

通过 DNA 算法自组装演示基本函数
具有计算功能的面向目标的元胞自动机是与特定规则相关的 DNA 算法自组装领域的主要挑战。我们通过使用可以实现到 DNA 构建块中的特定逻辑门来研究将元胞自动机原理用于数学学科的可行性。在这里,我们连接以下五个有代表性的初等函数: (i) 2、3、4 的倍数的枚举(通过3 输入/1 输出逻辑规则中的 R094、R062 和 R190证明);(ii) 0 和 1 的余数 (R132);(iii) 2 (R129) 的幂;(iv) n /2 和n 的上限函数/4(R152 和 R144);(v) 类似于由特定规则形成的 DNA 算法模式的湮灭模式 (R184)。在设计抽象积木和模拟算法格的生成后,我们进行了如下实验:设计具有特定粘性末端的 DNA 瓦片,通过两步退火方法,并使用原子力显微镜 (AFM) 验证给定 DNA 晶格上的预期算法模式。我们在给定的算法点阵上观察到了代表性的模式,例如水平和对角条纹以及嵌入的三角形。单个规则的平均错误率在 8.8% (R184) 到 11.9% (R062) 之间,所有规则的平均错误率为 10.6%。对通过 DNA 算法模式证明的基本函数的解释可以扩展到更复杂的函数,这可能会为实现具有实验获得的模式的函数的最终答案提供新的见解。
更新日期:2021-11-23
中文翻译:

通过 DNA 算法自组装演示基本函数
具有计算功能的面向目标的元胞自动机是与特定规则相关的 DNA 算法自组装领域的主要挑战。我们通过使用可以实现到 DNA 构建块中的特定逻辑门来研究将元胞自动机原理用于数学学科的可行性。在这里,我们连接以下五个有代表性的初等函数: (i) 2、3、4 的倍数的枚举(通过3 输入/1 输出逻辑规则中的 R094、R062 和 R190证明);(ii) 0 和 1 的余数 (R132);(iii) 2 (R129) 的幂;(iv) n /2 和n 的上限函数/4(R152 和 R144);(v) 类似于由特定规则形成的 DNA 算法模式的湮灭模式 (R184)。在设计抽象积木和模拟算法格的生成后,我们进行了如下实验:设计具有特定粘性末端的 DNA 瓦片,通过两步退火方法,并使用原子力显微镜 (AFM) 验证给定 DNA 晶格上的预期算法模式。我们在给定的算法点阵上观察到了代表性的模式,例如水平和对角条纹以及嵌入的三角形。单个规则的平均错误率在 8.8% (R184) 到 11.9% (R062) 之间,所有规则的平均错误率为 10.6%。对通过 DNA 算法模式证明的基本函数的解释可以扩展到更复杂的函数,这可能会为实现具有实验获得的模式的函数的最终答案提供新的见解。











































 京公网安备 11010802027423号
京公网安备 11010802027423号