当前位置:
X-MOL 学术
›
Comput. Struct. Biotechnol. J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Identification of the susceptibility genes for COVID-19 in lung adenocarcinoma with global data and biological computation methods
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-11-20 , DOI: 10.1016/j.csbj.2021.11.026 Li Gao 1 , Guo-Sheng Li 1 , Jian-Di Li 1 , Juan He 1 , Yu Zhang 2 , Hua-Fu Zhou 3 , Jin-Liang Kong 4 , Gang Chen 1
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-11-20 , DOI: 10.1016/j.csbj.2021.11.026 Li Gao 1 , Guo-Sheng Li 1 , Jian-Di Li 1 , Juan He 1 , Yu Zhang 2 , Hua-Fu Zhou 3 , Jin-Liang Kong 4 , Gang Chen 1
Affiliation
|
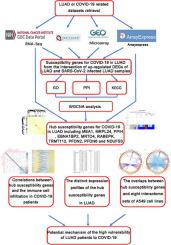
The risk of infection with COVID-19 is high in lung adenocarcinoma (LUAD) patients, and there is a dearth of studies on the molecular mechanism underlying the high susceptibility of LUAD patients to COVID-19 from the perspective of the global differential expression landscape. To fill the research void on the molecular mechanism underlying the high susceptibility of LUAD patients to COVID-19 from the perspective of the global differential expression landscape. Herein, we identified genes, specifically the differentially expressed genes (DEGs), correlated with the susceptibility of LUAD patients to COVID-19. These were obtained by calculating standard mean deviation (SMD) values for 49 SARS-CoV-2-infected LUAD samples and 24 non-affected LUAD samples, as well as 3931 LUAD samples and 3027 non-cancer lung samples from 40 pooled RNA-seq and microarray datasets. Hub susceptibility genes significantly related to COVID-19 were further selected by weighted gene co-expression network analysis. Then, the hub genes were further analyzed via an examination of their clinical significance in multiple datasets, a correlation analysis of the immune cell infiltration level, and their interactions with the interactome sets of the A549 cell line. A total of 257 susceptibility genes were identified, and these genes were associated with RNA splicing, mitochondrial functions, and proteasomes. Ten genes, MEA1, MRPL24, PPIH, EBNA1BP2, MRTO4, RABEPK, TRMT112, PFDN2, PFDN6, and NDUFS3, were confirmed to be the hub susceptibility genes for COVID-19 in LUAD patients, and the hub susceptibility genes were significantly correlated with the infiltration of multiple immune cells. In conclusion, the susceptibility genes for COVID-19 in LUAD patients discovered in this study may increase our understanding of the high risk of COVID-19 in LUAD patients.
中文翻译:

利用全局数据和生物计算方法鉴定肺腺癌中COVID-19的易感基因
肺腺癌(LUAD)患者感染COVID-19的风险较高,但从全球差异表达格局的角度对LUAD患者对COVID-19高易感性的分子机制缺乏研究。从全球差异表达格局的角度填补LUAD患者对COVID-19高易感性分子机制的研究空白。在此,我们鉴定了与 LUAD 患者对 COVID-19 易感性相关的基因,特别是差异表达基因 (DEG)。这些数据是通过计算 49 个 SARS-CoV-2 感染的 LUAD 样本和 24 个未受影响的 LUAD 样本,以及来自 40 个合并 RNA-seq 的 3931 个 LUAD 样本和 3027 个非癌症肺样本的标准均差 (SMD) 值而获得的。和微阵列数据集。通过加权基因共表达网络分析进一步筛选出与COVID-19显着相关的Hub易感基因。然后,通过检查其在多个数据集中的临床意义、免疫细胞浸润水平的相关性分析以及它们与 A549 细胞系的相互作用组集的相互作用,进一步分析了中心基因。总共鉴定出 257 个易感基因,这些基因与 RNA 剪接、线粒体功能和蛋白酶体相关。 MEA1、MRPL24、PPIH、EBNA1BP2、MRTO4、RABEPK、TRMT112、PFDN2、PFDN6和NDUFS3 10个基因被确认为LUAD患者中COVID-19的中枢易感基因,且中枢易感基因与多种免疫细胞浸润。 总之,本研究中发现的 LUAD 患者中的 COVID-19 易感基因可能会增加我们对 LUAD 患者中 COVID-19 高风险的认识。
更新日期:2021-11-20
中文翻译:

利用全局数据和生物计算方法鉴定肺腺癌中COVID-19的易感基因
肺腺癌(LUAD)患者感染COVID-19的风险较高,但从全球差异表达格局的角度对LUAD患者对COVID-19高易感性的分子机制缺乏研究。从全球差异表达格局的角度填补LUAD患者对COVID-19高易感性分子机制的研究空白。在此,我们鉴定了与 LUAD 患者对 COVID-19 易感性相关的基因,特别是差异表达基因 (DEG)。这些数据是通过计算 49 个 SARS-CoV-2 感染的 LUAD 样本和 24 个未受影响的 LUAD 样本,以及来自 40 个合并 RNA-seq 的 3931 个 LUAD 样本和 3027 个非癌症肺样本的标准均差 (SMD) 值而获得的。和微阵列数据集。通过加权基因共表达网络分析进一步筛选出与COVID-19显着相关的Hub易感基因。然后,通过检查其在多个数据集中的临床意义、免疫细胞浸润水平的相关性分析以及它们与 A549 细胞系的相互作用组集的相互作用,进一步分析了中心基因。总共鉴定出 257 个易感基因,这些基因与 RNA 剪接、线粒体功能和蛋白酶体相关。 MEA1、MRPL24、PPIH、EBNA1BP2、MRTO4、RABEPK、TRMT112、PFDN2、PFDN6和NDUFS3 10个基因被确认为LUAD患者中COVID-19的中枢易感基因,且中枢易感基因与多种免疫细胞浸润。 总之,本研究中发现的 LUAD 患者中的 COVID-19 易感基因可能会增加我们对 LUAD 患者中 COVID-19 高风险的认识。











































 京公网安备 11010802027423号
京公网安备 11010802027423号