当前位置:
X-MOL 学术
›
Comput. Struct. Biotechnol. J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
DeepFoci: Deep learning-based algorithm for fast automatic analysis of DNA double-strand break ionizing radiation-induced foci
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-11-18 , DOI: 10.1016/j.csbj.2021.11.019 Tomas Vicar 1, 2, 3 , Jaromir Gumulec 3 , Radim Kolar 1 , Olga Kopecna 2 , Eva Pagacova 2 , Iva Falkova 2 , Martin Falk 2
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-11-18 , DOI: 10.1016/j.csbj.2021.11.019 Tomas Vicar 1, 2, 3 , Jaromir Gumulec 3 , Radim Kolar 1 , Olga Kopecna 2 , Eva Pagacova 2 , Iva Falkova 2 , Martin Falk 2
Affiliation
|
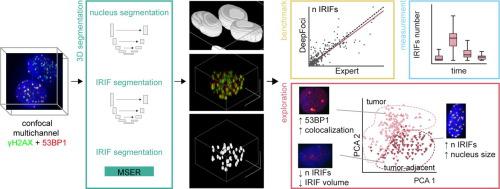
DNA double-strand breaks (DSBs), marked by ionizing radiation-induced (repair) foci (IRIFs), are the most serious DNA lesions and are dangerous to human health. IRIF quantification based on confocal microscopy represents the most sensitive and gold-standard method in radiation biodosimetry and allows research on DSB induction and repair at the molecular and single-cell levels. In this study, we introduce DeepFoci – a deep learning-based fully automatic method for IRIF counting and morphometric analysis. DeepFoci is designed to work with 3D multichannel data (trained for 53BP1 and γH2AX) and uses U-Net for nucleus segmentation and IRIF detection, together with maximally stable extremal region-based IRIF segmentation.
中文翻译:

DeepFoci:基于深度学习的算法,用于快速自动分析 DNA 双链断裂电离辐射引起的焦点
以电离辐射诱导(修复)灶(IRIF)为标志的 DNA 双链断裂(DSB)是最严重的 DNA 损伤,对人类健康有害。基于共焦显微镜的 IRIF 定量代表了辐射生物剂量测定中最灵敏和金标准的方法,并允许在分子和单细胞水平上研究 DSB 诱导和修复。在本研究中,我们介绍了 DeepFoci——一种基于深度学习的全自动 IRIF 计数和形态分析方法。 DeepFoci 设计用于处理 3D 多通道数据(针对 53BP1 和 γH2AX 进行训练),并使用 U-Net 进行核分割和 IRIF 检测,以及基于最稳定极值区域的 IRIF 分割。
更新日期:2021-11-18
中文翻译:

DeepFoci:基于深度学习的算法,用于快速自动分析 DNA 双链断裂电离辐射引起的焦点
以电离辐射诱导(修复)灶(IRIF)为标志的 DNA 双链断裂(DSB)是最严重的 DNA 损伤,对人类健康有害。基于共焦显微镜的 IRIF 定量代表了辐射生物剂量测定中最灵敏和金标准的方法,并允许在分子和单细胞水平上研究 DSB 诱导和修复。在本研究中,我们介绍了 DeepFoci——一种基于深度学习的全自动 IRIF 计数和形态分析方法。 DeepFoci 设计用于处理 3D 多通道数据(针对 53BP1 和 γH2AX 进行训练),并使用 U-Net 进行核分割和 IRIF 检测,以及基于最稳定极值区域的 IRIF 分割。











































 京公网安备 11010802027423号
京公网安备 11010802027423号