当前位置:
X-MOL 学术
›
Comput. Struct. Biotechnol. J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A comprehensive transcription factor and DNA-binding motif resource for the construction of gene regulatory networks in Botrytis cinerea and Trichoderma atroviride
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-11-18 , DOI: 10.1016/j.csbj.2021.11.012 Consuelo Olivares-Yañez 1, 2 , Evelyn Sánchez 1, 3 , Gabriel Pérez-Lara 1, 2 , Aldo Seguel 1, 4 , Pamela Y Camejo 1 , Luis F Larrondo 1, 4 , Elena A Vidal 1, 3, 5 , Paulo Canessa 1, 2
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-11-18 , DOI: 10.1016/j.csbj.2021.11.012 Consuelo Olivares-Yañez 1, 2 , Evelyn Sánchez 1, 3 , Gabriel Pérez-Lara 1, 2 , Aldo Seguel 1, 4 , Pamela Y Camejo 1 , Luis F Larrondo 1, 4 , Elena A Vidal 1, 3, 5 , Paulo Canessa 1, 2
Affiliation
|
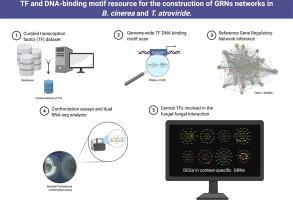
and are two relevant fungi in agricultural systems. To gain insights into these organisms’ transcriptional gene regulatory networks (GRNs), we generated a manually curated transcription factor (TF) dataset for each of them, followed by a GRN inference utilizing available sequence motifs describing DNA-binding specificity and global gene expression data. As a proof of concept of the usefulness of this resource to pinpoint key transcriptional regulators, we employed publicly available transcriptomics data and a newly generated dual RNA-seq dataset to build context-specific and GRNs under two different biological paradigms: exposure to continuous light and confrontation assays. Network analysis of fungal responses to constant light revealed striking differences in the transcriptional landscape of both fungi. On the other hand, we found that the confrontation of both microorganisms elicited a distinct set of differentially expressed genes with changes in exceeding those in . Using our regulatory network data, we were able to determine, in both fungi, central TFs involved in this interaction response, including TFs controlling a large set of extracellular peptidases in the biocontrol agent . In summary, our work provides a comprehensive catalog of transcription factors and regulatory interactions for both organisms. This catalog can now serve as a basis for generating novel hypotheses on transcriptional regulatory circuits in different experimental contexts.
中文翻译:

用于构建灰葡萄孢和深绿木霉基因调控网络的综合转录因子和 DNA 结合基序资源
和 是农业系统中的两种相关真菌。为了深入了解这些生物体的转录基因调控网络 (GRN),我们为每个生物体生成了手动整理的转录因子 (TF) 数据集,然后利用描述 DNA 结合特异性的可用序列基序和全局基因表达数据进行 GRN 推断。作为该资源在查明关键转录调节因子方面的有用性的概念证明,我们采用了公开的转录组学数据和新生成的双 RNA-seq 数据集,在两种不同的生物范式下构建上下文特定的 GRN:暴露于连续光和对抗分析。真菌对恒定光反应的网络分析揭示了两种真菌转录景观的显着差异。另一方面,我们发现两种微生物的对抗引发了一组独特的差异表达基因,其变化超过了 .利用我们的调控网络数据,我们能够确定两种真菌中参与这种相互作用反应的中心转录因子,包括控制生物防治剂中大量胞外肽酶的转录因子。总之,我们的工作提供了两种生物体的转录因子和调控相互作用的全面目录。该目录现在可以作为在不同实验背景下生成关于转录调节回路的新假设的基础。
更新日期:2021-11-18
中文翻译:

用于构建灰葡萄孢和深绿木霉基因调控网络的综合转录因子和 DNA 结合基序资源
和 是农业系统中的两种相关真菌。为了深入了解这些生物体的转录基因调控网络 (GRN),我们为每个生物体生成了手动整理的转录因子 (TF) 数据集,然后利用描述 DNA 结合特异性的可用序列基序和全局基因表达数据进行 GRN 推断。作为该资源在查明关键转录调节因子方面的有用性的概念证明,我们采用了公开的转录组学数据和新生成的双 RNA-seq 数据集,在两种不同的生物范式下构建上下文特定的 GRN:暴露于连续光和对抗分析。真菌对恒定光反应的网络分析揭示了两种真菌转录景观的显着差异。另一方面,我们发现两种微生物的对抗引发了一组独特的差异表达基因,其变化超过了 .利用我们的调控网络数据,我们能够确定两种真菌中参与这种相互作用反应的中心转录因子,包括控制生物防治剂中大量胞外肽酶的转录因子。总之,我们的工作提供了两种生物体的转录因子和调控相互作用的全面目录。该目录现在可以作为在不同实验背景下生成关于转录调节回路的新假设的基础。











































 京公网安备 11010802027423号
京公网安备 11010802027423号