当前位置:
X-MOL 学术
›
Comput. Struct. Biotechnol. J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Atomic-scale insights into allosteric inhibition and evolutional rescue mechanism of Streptococcus thermophilus Cas9 by the anti-CRISPR protein AcrIIA6
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-11-16 , DOI: 10.1016/j.csbj.2021.11.010 Xinyi Li 1, 2 , Chengxiang Wang 2 , Ting Peng 2 , Zongtao Chai 3 , Duan Ni 4 , Yaqin Liu 5 , Jian Zhang 2, 5 , Ting Chen 1 , Shaoyong Lu 2, 5
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-11-16 , DOI: 10.1016/j.csbj.2021.11.010 Xinyi Li 1, 2 , Chengxiang Wang 2 , Ting Peng 2 , Zongtao Chai 3 , Duan Ni 4 , Yaqin Liu 5 , Jian Zhang 2, 5 , Ting Chen 1 , Shaoyong Lu 2, 5
Affiliation
|
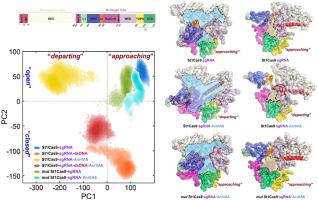
CRISPR-Cas systems are prokaryotic adaptive immunity against invading phages and plasmids. Phages have evolved diverse protein inhibitors of CRISPR-Cas systems, called anti-CRISPR (Acr) proteins, to neutralize this CRISPR machinery. In response, bacteria have co-evolved Cas variants to escape phage’s anti-CRISPR strategies, called anti-anti-CRISPR systems. Here we explore the anti-CRISPR allosteric inhibition and anti-anti-CRISPR rescue mechanisms between Cas9 (St1Cas9) and the anti-CRISPR protein AcrIIA6 at the atomic level, by generating mutants of key residues in St1Cas9. Extensive unbiased molecular dynamics simulations show that the functional motions of St1Cas9 in the presence of AcrIIA6 differ substantially from those of St1Cas9 alone. AcrIIA6 binding triggers a shift of St1Cas9 conformational ensemble towards a less catalytically competent state; this state significantly compromises protospacer adjacent motif (PAM) recognition and nuclease activity by altering interdependently conformational dynamics and allosteric signals among nuclease domains, PAM-interacting (PI) regions, and AcrIIA6 binding motifs. Via DNA cleavage assays, we further elucidate the rescue mechanism of efficiently escaping AcrIIA6 inhibition harboring St1Cas9 triple mutations (G993K/K1008M/K1010E) in the PI domain and identify the evolutionary landscape of such mutational escape within species. Our results provide mechanistic insights into Acr proteins as natural brakes for the CRISPR-Cas systems and a promising potential for the design of allosteric Acr peptidomimetics.
中文翻译:

抗 CRISPR 蛋白 AcrIIA6 对嗜热链球菌 Cas9 的变构抑制和进化拯救机制的原子尺度见解
CRISPR-Cas系统是针对入侵噬菌体和质粒的原核适应性免疫。噬菌体进化出了多种 CRISPR-Cas 系统的蛋白质抑制剂,称为抗 CRISPR (Acr) 蛋白质,以中和这种 CRISPR 机制。作为回应,细菌共同进化出 Cas 变体来逃避噬菌体的抗 CRISPR 策略,称为抗抗 CRISPR 系统。在这里,我们通过生成 St1Cas9 中关键残基的突变体,在原子水平上探索 Cas9 (St1Cas9) 和抗 CRISPR 蛋白 AcrIIA6 之间的抗 CRISPR 变构抑制和抗 CRISPR 救援机制。广泛的无偏分子动力学模拟表明,AcrIIA6 存在下 St1Cas9 的功能运动与单独 St1Cas9 的功能运动有很大不同。 AcrIIA6 结合触发 St1Cas9 构象整体向催化能力较低的状态转变;这种状态通过改变核酸酶结构域、PAM 相互作用 (PI) 区域和 AcrIIA6 结合基序之间相互依赖的构象动力学和变构信号,显着损害原型间隔子相邻基序 (PAM) 识别和核酸酶活性。通过DNA切割测定,我们进一步阐明了有效逃避PI结构域中含有St1Cas9三重突变(G993K/K1008M/K1010E)的AcrIIA6抑制的救援机制,并确定了物种内此类突变逃逸的进化景观。我们的结果为 Acr 蛋白作为 CRISPR-Cas 系统的天然制动器提供了机制见解,并为变构 Acr 肽模拟物的设计提供了广阔的前景。
更新日期:2021-11-16
中文翻译:

抗 CRISPR 蛋白 AcrIIA6 对嗜热链球菌 Cas9 的变构抑制和进化拯救机制的原子尺度见解
CRISPR-Cas系统是针对入侵噬菌体和质粒的原核适应性免疫。噬菌体进化出了多种 CRISPR-Cas 系统的蛋白质抑制剂,称为抗 CRISPR (Acr) 蛋白质,以中和这种 CRISPR 机制。作为回应,细菌共同进化出 Cas 变体来逃避噬菌体的抗 CRISPR 策略,称为抗抗 CRISPR 系统。在这里,我们通过生成 St1Cas9 中关键残基的突变体,在原子水平上探索 Cas9 (St1Cas9) 和抗 CRISPR 蛋白 AcrIIA6 之间的抗 CRISPR 变构抑制和抗 CRISPR 救援机制。广泛的无偏分子动力学模拟表明,AcrIIA6 存在下 St1Cas9 的功能运动与单独 St1Cas9 的功能运动有很大不同。 AcrIIA6 结合触发 St1Cas9 构象整体向催化能力较低的状态转变;这种状态通过改变核酸酶结构域、PAM 相互作用 (PI) 区域和 AcrIIA6 结合基序之间相互依赖的构象动力学和变构信号,显着损害原型间隔子相邻基序 (PAM) 识别和核酸酶活性。通过DNA切割测定,我们进一步阐明了有效逃避PI结构域中含有St1Cas9三重突变(G993K/K1008M/K1010E)的AcrIIA6抑制的救援机制,并确定了物种内此类突变逃逸的进化景观。我们的结果为 Acr 蛋白作为 CRISPR-Cas 系统的天然制动器提供了机制见解,并为变构 Acr 肽模拟物的设计提供了广阔的前景。











































 京公网安备 11010802027423号
京公网安备 11010802027423号