当前位置:
X-MOL 学术
›
Acc. Chem. Res.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Therapeutic mRNA Engineering from Head to Tail
Accounts of Chemical Research ( IF 16.4 ) Pub Date : 2021-11-10 , DOI: 10.1021/acs.accounts.1c00541 Longfei Jia 1 , Shu-Bing Qian 1
Accounts of Chemical Research ( IF 16.4 ) Pub Date : 2021-11-10 , DOI: 10.1021/acs.accounts.1c00541 Longfei Jia 1 , Shu-Bing Qian 1
Affiliation
|
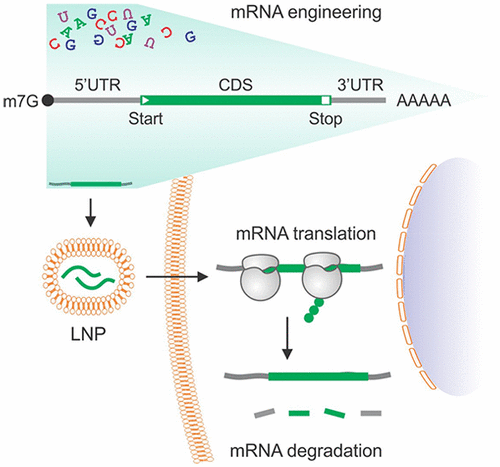
Synthetic messenger RNA (mRNA), once delivered into cells, can be readily translated into proteins by ribosomes, which do not distinguish exogenous mRNAs from endogenous transcripts. Until recently, the intrinsic instability and immunostimulatory property of exogenous RNAs largely hindered the therapeutic application of synthetic mRNAs. Thanks to major technological innovations, such as introduction of chemically modified nucleosides, synthetic mRNAs have become programmable therapeutic reagents. Compared to DNA or protein-based therapeutic reagents, synthetic mRNAs bear several advantages: flexible design, easy optimization, low-cost preparation, and scalable synthesis. Therapeutic mRNAs are commonly designed to encode specific antigens to elicit organismal immune response to pathogens like viruses, express functional proteins to replace defective ones inside cells, or introduce novel enzymes to achieve unique functions like genome editing. Recent years have witnessed stunning progress on the development of mRNA vaccines against SARS-Cov2. This success is built upon our fundamental understanding of mRNA metabolism and translational control, a knowledge accumulated during the past several decades. Given the astronomical number of sequence combinations of four nucleotides, sequence-dependent control of mRNA translation remains incompletely understood. Rational design of synthetic mRNAs with robust translation and optimal stability remains challenging. Massively paralleled reporter assay (MPRA) has been proven to be powerful in identifying sequence elements in controlling mRNA translatability and stability. Indeed, a completely randomized sequence in 5′ untranslated region (5′UTR) drives a wide range of translational outputs. In this Account, we will discuss general principles of mRNA translation in eukaryotic cells and elucidate the role of coding and noncoding regions in the translational regulation. From the therapeutic perspective, we will highlight the unique features of 5′ cap, 5′UTR, coding region (CDS), stop codon, 3′UTR, and poly(A) tail. By focusing on the design strategies in mRNA engineering, we hope this Account will contribute to the rational design of synthetic mRNAs with broad therapeutic potential.
中文翻译:

从头到尾的治疗性 mRNA 工程
合成的信使 RNA (mRNA) 一旦被递送到细胞中,就可以很容易地被核糖体翻译成蛋白质,核糖体不区分外源性 mRNA 和内源性转录物。直到最近,外源 RNA 的内在不稳定性和免疫刺激特性在很大程度上阻碍了合成 mRNA 的治疗应用。由于重大的技术创新,例如引入化学修饰的核苷,合成的 mRNA 已成为可编程的治疗试剂。与基于 DNA 或蛋白质的治疗试剂相比,合成 mRNA 具有几个优点:设计灵活、易于优化、制备成本低和合成规模可扩展。治疗性 mRNA 通常设计用于编码特定抗原,以引发机体对病毒等病原体的免疫反应,表达功能性蛋白质以替代细胞内有缺陷的蛋白质,或引入新的酶以实现基因组编辑等独特功能。近年来,针对 SARS-Cov2 的 mRNA 疫苗的开发取得了惊人的进展。这一成功建立在我们对 mRNA 代谢和翻译控制的基本理解之上,这是过去几十年积累的知识。鉴于四个核苷酸的序列组合的天文数量,mRNA翻译的序列依赖性控制仍然不完全清楚。具有稳健翻译和最佳稳定性的合成 mRNA 的合理设计仍然具有挑战性。大规模平行报告基因分析 (MPRA) 已被证明在识别控制 mRNA 可翻译性和稳定性的序列元件方面非常强大。的确,5'非翻译区(5'UTR)中的完全随机序列驱动广泛的翻译输出。在本文中,我们将讨论真核细胞中 mRNA 翻译的一般原理,并阐明编码区和非编码区在翻译调控中的作用。从治疗的角度来看,我们将突出 5' cap、5'UTR、编码区 (CDS)、终止密码子、3'UTR 和 poly(A) 尾的独特特征。通过关注 mRNA 工程中的设计策略,我们希望该帐户将有助于合理设计具有广泛治疗潜力的合成 mRNA。我们将讨论真核细胞中 mRNA 翻译的一般原理,并阐明编码区和非编码区在翻译调控中的作用。从治疗的角度来看,我们将突出 5' cap、5'UTR、编码区 (CDS)、终止密码子、3'UTR 和 poly(A) 尾的独特特征。通过关注 mRNA 工程中的设计策略,我们希望该帐户将有助于合理设计具有广泛治疗潜力的合成 mRNA。我们将讨论真核细胞中 mRNA 翻译的一般原理,并阐明编码区和非编码区在翻译调控中的作用。从治疗的角度来看,我们将突出 5' cap、5'UTR、编码区 (CDS)、终止密码子、3'UTR 和 poly(A) 尾的独特特征。通过关注 mRNA 工程中的设计策略,我们希望该帐户将有助于合理设计具有广泛治疗潜力的合成 mRNA。
更新日期:2021-12-07
中文翻译:

从头到尾的治疗性 mRNA 工程
合成的信使 RNA (mRNA) 一旦被递送到细胞中,就可以很容易地被核糖体翻译成蛋白质,核糖体不区分外源性 mRNA 和内源性转录物。直到最近,外源 RNA 的内在不稳定性和免疫刺激特性在很大程度上阻碍了合成 mRNA 的治疗应用。由于重大的技术创新,例如引入化学修饰的核苷,合成的 mRNA 已成为可编程的治疗试剂。与基于 DNA 或蛋白质的治疗试剂相比,合成 mRNA 具有几个优点:设计灵活、易于优化、制备成本低和合成规模可扩展。治疗性 mRNA 通常设计用于编码特定抗原,以引发机体对病毒等病原体的免疫反应,表达功能性蛋白质以替代细胞内有缺陷的蛋白质,或引入新的酶以实现基因组编辑等独特功能。近年来,针对 SARS-Cov2 的 mRNA 疫苗的开发取得了惊人的进展。这一成功建立在我们对 mRNA 代谢和翻译控制的基本理解之上,这是过去几十年积累的知识。鉴于四个核苷酸的序列组合的天文数量,mRNA翻译的序列依赖性控制仍然不完全清楚。具有稳健翻译和最佳稳定性的合成 mRNA 的合理设计仍然具有挑战性。大规模平行报告基因分析 (MPRA) 已被证明在识别控制 mRNA 可翻译性和稳定性的序列元件方面非常强大。的确,5'非翻译区(5'UTR)中的完全随机序列驱动广泛的翻译输出。在本文中,我们将讨论真核细胞中 mRNA 翻译的一般原理,并阐明编码区和非编码区在翻译调控中的作用。从治疗的角度来看,我们将突出 5' cap、5'UTR、编码区 (CDS)、终止密码子、3'UTR 和 poly(A) 尾的独特特征。通过关注 mRNA 工程中的设计策略,我们希望该帐户将有助于合理设计具有广泛治疗潜力的合成 mRNA。我们将讨论真核细胞中 mRNA 翻译的一般原理,并阐明编码区和非编码区在翻译调控中的作用。从治疗的角度来看,我们将突出 5' cap、5'UTR、编码区 (CDS)、终止密码子、3'UTR 和 poly(A) 尾的独特特征。通过关注 mRNA 工程中的设计策略,我们希望该帐户将有助于合理设计具有广泛治疗潜力的合成 mRNA。我们将讨论真核细胞中 mRNA 翻译的一般原理,并阐明编码区和非编码区在翻译调控中的作用。从治疗的角度来看,我们将突出 5' cap、5'UTR、编码区 (CDS)、终止密码子、3'UTR 和 poly(A) 尾的独特特征。通过关注 mRNA 工程中的设计策略,我们希望该帐户将有助于合理设计具有广泛治疗潜力的合成 mRNA。











































 京公网安备 11010802027423号
京公网安备 11010802027423号