当前位置:
X-MOL 学术
›
Comput. Struct. Biotechnol. J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Bacterial species identification using MALDI-TOF mass spectrometry and machine learning techniques: A large-scale benchmarking study
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-11-09 , DOI: 10.1016/j.csbj.2021.11.004 Thomas Mortier 1 , Anneleen D Wieme 2 , Peter Vandamme 2 , Willem Waegeman 1
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-11-09 , DOI: 10.1016/j.csbj.2021.11.004 Thomas Mortier 1 , Anneleen D Wieme 2 , Peter Vandamme 2 , Willem Waegeman 1
Affiliation
|
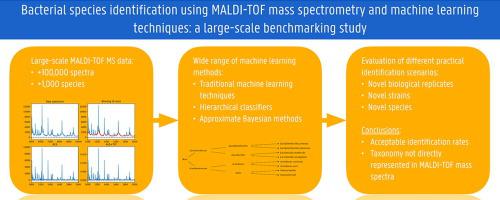
Today machine learning methods are commonly deployed for bacterial species identification using MALDI-TOF mass spectrometry data. However, most of the studies reported in literature only consider very traditional machine learning methods on small datasets that contain a limited number of species. In this paper we present benchmarking results on an unprecedented scale for a wide range of machine learning methods, using datasets that contain almost 100,000 spectra and more than 1000 different species. The size and the diversity of the data allow to compare three important identification scenarios that are often not distinguished in literature, i.e., identification for novel biological replicates, novel strains and novel species that are not present in the training data. The results demonstrate that in all three scenarios acceptable identification rates are obtained, but the numbers are typically lower than those reported in studies with a more limited analysis. Using hierarchical classification methods, we also demonstrate that taxonomic information is in general not well preserved in MALDI-TOF mass spectrometry data. For the novel species scenario, we apply for the first time neural networks with Monte Carlo dropout, which have shown to be successful in other domains, such as computer vision, for the detection of novel species.
中文翻译:

使用 MALDI-TOF 质谱和机器学习技术鉴定细菌物种:大规模基准研究
如今,机器学习方法通常用于使用 MALDI-TOF 质谱数据进行细菌物种鉴定。然而,文献中报道的大多数研究只考虑在包含有限数量物种的小数据集上使用非常传统的机器学习方法。在本文中,我们使用包含近 100,000 个光谱和 1000 多个不同物种的数据集,以前所未有的规模展示了各种机器学习方法的基准测试结果。数据的大小和多样性允许比较文献中通常未区分的三种重要识别场景,即识别训练数据中不存在的新生物重复、新菌株和新物种。结果表明,在所有三种情况下都获得了可接受的识别率,但该数字通常低于分析更有限的研究中报告的数字。使用分层分类方法,我们还证明分类信息通常在 MALDI-TOF 质谱数据中没有得到很好的保存。对于新物种场景,我们首次应用带有蒙特卡罗 dropout 的神经网络,该网络已被证明在其他领域(例如计算机视觉)成功检测新物种。
更新日期:2021-11-09
中文翻译:

使用 MALDI-TOF 质谱和机器学习技术鉴定细菌物种:大规模基准研究
如今,机器学习方法通常用于使用 MALDI-TOF 质谱数据进行细菌物种鉴定。然而,文献中报道的大多数研究只考虑在包含有限数量物种的小数据集上使用非常传统的机器学习方法。在本文中,我们使用包含近 100,000 个光谱和 1000 多个不同物种的数据集,以前所未有的规模展示了各种机器学习方法的基准测试结果。数据的大小和多样性允许比较文献中通常未区分的三种重要识别场景,即识别训练数据中不存在的新生物重复、新菌株和新物种。结果表明,在所有三种情况下都获得了可接受的识别率,但该数字通常低于分析更有限的研究中报告的数字。使用分层分类方法,我们还证明分类信息通常在 MALDI-TOF 质谱数据中没有得到很好的保存。对于新物种场景,我们首次应用带有蒙特卡罗 dropout 的神经网络,该网络已被证明在其他领域(例如计算机视觉)成功检测新物种。











































 京公网安备 11010802027423号
京公网安备 11010802027423号