当前位置:
X-MOL 学术
›
Comput. Struct. Biotechnol. J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Mitogenome-wide comparison and phylogeny reveal group I intron dynamics and intraspecific diversification within the phytopathogen Corynespora cassiicola
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-11-03 , DOI: 10.1016/j.csbj.2021.11.002 Qingzhou Ma 1 , Haiyan Wu 2 , Yuehua Geng 1 , Qiang Li 3 , Rui Zang 1 , Yashuang Guo 1 , Chao Xu 1 , Meng Zhang 1
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-11-03 , DOI: 10.1016/j.csbj.2021.11.002 Qingzhou Ma 1 , Haiyan Wu 2 , Yuehua Geng 1 , Qiang Li 3 , Rui Zang 1 , Yashuang Guo 1 , Chao Xu 1 , Meng Zhang 1
Affiliation
|
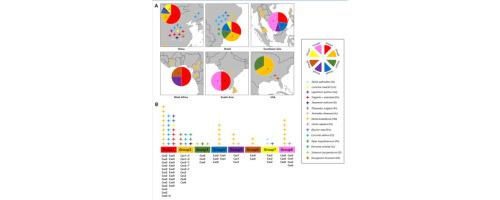
, the causal agent of an extensive range of plant diseases worldwide, is a momentous fungus with diverse lifestyles and rich in intraspecies variations. In the present study, a total of 56 mitochondrial genomes of were assembled (except two available online) and analyzed, of which 16 mitogenomes were newly sequenced here. All these circular mitochondrial DNA (mtDNA) molecules, ranging from 39,223 bp to 45,786 bp in length, comprised the same set of 13 core protein-coding genes (PCGs), two rRNAs and 27 tRNAs arranged in identical order. Across the above conserved genes, had the largest genetic distance between different isolates and was possibly subjected to positive selection pressure. Comparative mitogenomic analysis indicated that seven group I (IB, IC1, and IC2) introns with a length range of 1013–1876 bp were differentially inserted in three core PCGs (, , and ), resulting in the varied mitogenome sizes among isolates. In combination with dynamic distribution of the introns, a well-supported mitogenome-wide phylogeny of the 56 isolates revealed eight phylogenetic groups, which only had weak correlations with host range and toxin class. Different groups of isolates exhibited obvious differences in length and GC content of some genes, while a degree of variance in codon usage and tRNA structure was also observed. This research served as the first report on mitogenomic comparisons within , and could provide new insights into its intraspecific microevolution and genetic diversity.
中文翻译:

全线粒体基因组比较和系统发育揭示了植物病原体 Corynespora cassiicola 内的 I 组内含子动态和种内多样性
真菌是世界范围内多种植物病害的病原体,是一种重要的真菌,具有多种生活方式和丰富的种内变异。在本研究中,总共组装并分析了 56 个线粒体基因组(除了两个在线可用的基因组),其中 16 个线粒体基因组是这里新测序的。所有这些环状线粒体 DNA (mtDNA) 分子的长度从 39,223 bp 到 45,786 bp 不等,包含同一组 13 个核心蛋白编码基因 (PCG)、两个 rRNA 和 27 个 tRNA,它们以相同的顺序排列。在上述保守基因中,不同分离株之间的遗传距离最大,可能受到正选择压力。比较线粒体基因组分析表明,长度范围为 1013-1876 bp 的 7 个 I 组(IB、IC1 和 IC2)内含子差异插入到三个核心 PCG(、、和)中,导致分离株之间的线粒体基因组大小不同。结合内含子的动态分布,56 个分离株的全线粒体系统发育得到充分支持,揭示了 8 个系统发育组,这些组与宿主范围和毒素类别仅具有弱相关性。不同组的分离株在一些基因的长度和GC含量上表现出明显的差异,同时在密码子使用和tRNA结构上也观察到一定程度的差异。这项研究是第一份关于线粒体基因组比较的报告,可以为其种内微进化和遗传多样性提供新的见解。
更新日期:2021-11-03
中文翻译:

全线粒体基因组比较和系统发育揭示了植物病原体 Corynespora cassiicola 内的 I 组内含子动态和种内多样性
真菌是世界范围内多种植物病害的病原体,是一种重要的真菌,具有多种生活方式和丰富的种内变异。在本研究中,总共组装并分析了 56 个线粒体基因组(除了两个在线可用的基因组),其中 16 个线粒体基因组是这里新测序的。所有这些环状线粒体 DNA (mtDNA) 分子的长度从 39,223 bp 到 45,786 bp 不等,包含同一组 13 个核心蛋白编码基因 (PCG)、两个 rRNA 和 27 个 tRNA,它们以相同的顺序排列。在上述保守基因中,不同分离株之间的遗传距离最大,可能受到正选择压力。比较线粒体基因组分析表明,长度范围为 1013-1876 bp 的 7 个 I 组(IB、IC1 和 IC2)内含子差异插入到三个核心 PCG(、、和)中,导致分离株之间的线粒体基因组大小不同。结合内含子的动态分布,56 个分离株的全线粒体系统发育得到充分支持,揭示了 8 个系统发育组,这些组与宿主范围和毒素类别仅具有弱相关性。不同组的分离株在一些基因的长度和GC含量上表现出明显的差异,同时在密码子使用和tRNA结构上也观察到一定程度的差异。这项研究是第一份关于线粒体基因组比较的报告,可以为其种内微进化和遗传多样性提供新的见解。











































 京公网安备 11010802027423号
京公网安备 11010802027423号