当前位置:
X-MOL 学术
›
Comput. Struct. Biotechnol. J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Is there an advantageous arrangement of aromatic residues in proteins? Statistical analysis of aromatic interactions in globular proteins
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-11-01 , DOI: 10.1016/j.csbj.2021.10.036 Mikhail Yu Lobanov 1 , Leonid B Pereyaslavets 1 , Ilya V Likhachev 1, 2 , Bakhyt T Matkarimov 3 , Oxana V Galzitskaya 1, 4
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-11-01 , DOI: 10.1016/j.csbj.2021.10.036 Mikhail Yu Lobanov 1 , Leonid B Pereyaslavets 1 , Ilya V Likhachev 1, 2 , Bakhyt T Matkarimov 3 , Oxana V Galzitskaya 1, 4
Affiliation
|
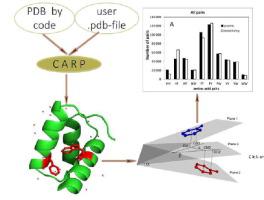
The aim of this study was to evaluate the favorability of different conformations of aromatic residues in proteins by analysing the occurrence of particular conformations. The clustering of protein structures from the Protein Data Bank (PDB) was performed. Conformations of interacting aromatic residues were analyzed for 511282 pairs in 35493 protein structures sharing less than 50% identity. Pairs with a parallel arrangement of aromatic residues made up 6.2% of all possible ones, which was twice as much as expected. Pairs with a perpendicular arrangement of aromatic residues made up 25%. We demonstrate that the most favorable arrangement was at an angle of 60° between the interacting aromatic residues. Among all possible aromatic pairs, the His-His pair was twice as frequent as expected, and the His-Phe pair was less frequent than expected. A server (CARP – Contacts of Aromatic Residues in Proteins) has been created for calculating essential structural features of interacting aromatic residues in proteins: .
中文翻译:

蛋白质中芳香族残基是否存在有利的排列?球状蛋白芳香族相互作用的统计分析
本研究的目的是通过分析特定构象的出现来评估蛋白质中芳香残基不同构象的有利性。对蛋白质数据库 (PDB) 中的蛋白质结构进行聚类。对 35493 个蛋白质结构中的 511282 对相互作用的芳香族残基的构象进行了分析,这些残基的同一性低于 50%。芳香族残基平行排列的配对占所有可能配对的 6.2%,是预期的两倍。芳香族残基垂直排列的对占25%。我们证明最有利的排列是相互作用的芳香族残基之间呈 60° 角。在所有可能的芳香族对中,His-His 对的出现频率是预期的两倍,而 His-Phe 对的出现频率低于预期。创建了一个服务器(CARP – 蛋白质中芳香残基的接触),用于计算蛋白质中相互作用的芳香残基的基本结构特征:。
更新日期:2021-11-01
中文翻译:

蛋白质中芳香族残基是否存在有利的排列?球状蛋白芳香族相互作用的统计分析
本研究的目的是通过分析特定构象的出现来评估蛋白质中芳香残基不同构象的有利性。对蛋白质数据库 (PDB) 中的蛋白质结构进行聚类。对 35493 个蛋白质结构中的 511282 对相互作用的芳香族残基的构象进行了分析,这些残基的同一性低于 50%。芳香族残基平行排列的配对占所有可能配对的 6.2%,是预期的两倍。芳香族残基垂直排列的对占25%。我们证明最有利的排列是相互作用的芳香族残基之间呈 60° 角。在所有可能的芳香族对中,His-His 对的出现频率是预期的两倍,而 His-Phe 对的出现频率低于预期。创建了一个服务器(CARP – 蛋白质中芳香残基的接触),用于计算蛋白质中相互作用的芳香残基的基本结构特征:。











































 京公网安备 11010802027423号
京公网安备 11010802027423号