当前位置:
X-MOL 学术
›
Chem. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Deep generative design with 3D pharmacophoric constraints
Chemical Science ( IF 7.6 ) Pub Date : 2021-10-25 , DOI: 10.1039/d1sc02436a Fergus Imrie 1 , Thomas E Hadfield 1 , Anthony R Bradley 2 , Charlotte M Deane 1
Chemical Science ( IF 7.6 ) Pub Date : 2021-10-25 , DOI: 10.1039/d1sc02436a Fergus Imrie 1 , Thomas E Hadfield 1 , Anthony R Bradley 2 , Charlotte M Deane 1
Affiliation
|
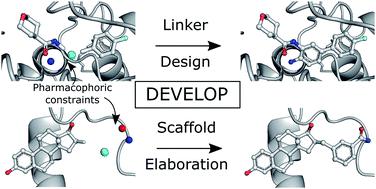
Generative models have increasingly been proposed as a solution to the molecular design problem. However, it has proved challenging to control the design process or incorporate prior knowledge, limiting their practical use in drug discovery. In particular, generative methods have made limited use of three-dimensional (3D) structural information even though this is critical to binding. This work describes a method to incorporate such information and demonstrates the benefit of doing so. We combine an existing graph-based deep generative model, DeLinker, with a convolutional neural network to utilise physically-meaningful 3D representations of molecules and target pharmacophores. We apply our model, DEVELOP, to both linker and R-group design, demonstrating its suitability for both hit-to-lead and lead optimisation. The 3D pharmacophoric information results in improved generation and allows greater control of the design process. In multiple large-scale evaluations, we show that including 3D pharmacophoric constraints results in substantial improvements in the quality of generated molecules. On a challenging test set derived from PDBbind, our model improves the proportion of generated molecules with high 3D similarity to the original molecule by over 300%. In addition, DEVELOP recovers 10× more of the original molecules compared to the baseline DeLinker method. Our approach is general-purpose, readily modifiable to alternate 3D representations, and can be incorporated into other generative frameworks. Code is available at https://github.com/oxpig/DEVELOP.
中文翻译:

具有 3D 药效团约束的深度生成设计
生成模型越来越多地被提出作为分子设计问题的解决方案。然而,事实证明,控制设计过程或整合先验知识具有挑战性,限制了它们在药物发现中的实际应用。特别是,生成方法对三维 (3D) 结构信息的使用有限,尽管这对于绑定至关重要。这项工作描述了一种合并此类信息的方法,并展示了这样做的好处。我们将现有的基于图的深度生成模型 DeLinker 与卷积神经网络相结合,以利用分子和目标药效团的物理意义的 3D 表示。我们将我们的模型 DEVELOP 应用于连接子和 R 基团设计,证明其适用于从命中到先导和先导优化。 3D 药效团信息可改进生成并允许更好地控制设计过程。在多个大规模评估中,我们表明,包含 3D 药效团约束可以显着提高生成分子的质量。在源自 PDBbind 的具有挑战性的测试集上,我们的模型将生成的与原始分子具有高 3D 相似性的分子的比例提高了 300% 以上。此外,与基线 DeLinker 方法相比,DEVELOP 回收的原始分子多了 10 倍。我们的方法是通用的,很容易修改为替代 3D 表示,并且可以合并到其他生成框架中。代码可在 https://github.com/oxpig/DEVELOP 获取。
更新日期:2021-10-26
中文翻译:

具有 3D 药效团约束的深度生成设计
生成模型越来越多地被提出作为分子设计问题的解决方案。然而,事实证明,控制设计过程或整合先验知识具有挑战性,限制了它们在药物发现中的实际应用。特别是,生成方法对三维 (3D) 结构信息的使用有限,尽管这对于绑定至关重要。这项工作描述了一种合并此类信息的方法,并展示了这样做的好处。我们将现有的基于图的深度生成模型 DeLinker 与卷积神经网络相结合,以利用分子和目标药效团的物理意义的 3D 表示。我们将我们的模型 DEVELOP 应用于连接子和 R 基团设计,证明其适用于从命中到先导和先导优化。 3D 药效团信息可改进生成并允许更好地控制设计过程。在多个大规模评估中,我们表明,包含 3D 药效团约束可以显着提高生成分子的质量。在源自 PDBbind 的具有挑战性的测试集上,我们的模型将生成的与原始分子具有高 3D 相似性的分子的比例提高了 300% 以上。此外,与基线 DeLinker 方法相比,DEVELOP 回收的原始分子多了 10 倍。我们的方法是通用的,很容易修改为替代 3D 表示,并且可以合并到其他生成框架中。代码可在 https://github.com/oxpig/DEVELOP 获取。











































 京公网安备 11010802027423号
京公网安备 11010802027423号