Protein & Cell ( IF 13.6 ) Pub Date : 2021-10-22 , DOI: 10.1007/s13238-021-00885-0 Feisheng Zhong 1, 2 , Xiaolong Wu 1, 3 , Ruirui Yang 1, 2, 4 , Xutong Li 1, 2 , Dingyan Wang 1, 2 , Zunyun Fu 1, 5 , Xiaohong Liu 1, 4 , XiaoZhe Wan 1, 2 , Tianbiao Yang 1, 2 , Zisheng Fan 1, 5 , Yinghui Zhang 1, 2 , Xiaomin Luo 1, 2 , Kaixian Chen 1, 2 , Sulin Zhang 1, 2 , Hualiang Jiang 1, 2, 3, 4 , Mingyue Zheng 1, 2, 5
|
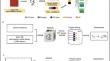
A fundamental challenge that arises in biomedicine is the need to characterize compounds in a relevant cellular context in order to reveal potential on-target or off-target effects. Recently, the fast accumulation of gene transcriptional profiling data provides us an unprecedented opportunity to explore the protein targets of chemical compounds from the perspective of cell transcriptomics and RNA biology. Here, we propose a novel Siamese spectral-based graph convolutional network (SSGCN) model for inferring the protein targets of chemical compounds from gene transcriptional profiles. Although the gene signature of a compound perturbation only provides indirect clues of the interacting targets, and the biological networks under different experiment conditions further complicate the situation, the SSGCN model was successfully trained to learn from known compound-target pairs by uncovering the hidden correlations between compound perturbation profiles and gene knockdown profiles. On a benchmark set and a large time-split validation dataset, the model achieved higher target inference accuracy as compared to previous methods such as Connectivity Map. Further experimental validations of prediction results highlight the practical usefulness of SSGCN in either inferring the interacting targets of compound, or reversely, in finding novel inhibitors of a given target of interest.
中文翻译:

使用新颖的图卷积网络框架通过挖掘转录数据来推断药物靶标
生物医学中出现的一个基本挑战是需要在相关细胞环境中表征化合物,以揭示潜在的靶向或脱靶效应。近年来,基因转录谱数据的快速积累为我们从细胞转录组学和RNA生物学的角度探索化合物的蛋白质靶标提供了前所未有的机会。在这里,我们提出了一种新颖的基于暹罗谱的图卷积网络(SSGCN)模型,用于从基因转录谱中推断化合物的蛋白质靶标。尽管复合扰动的基因特征仅提供相互作用目标的间接线索,并且不同实验条件下的生物网络使情况进一步复杂化,但 SSGCN 模型已成功训练为通过揭示之间隐藏的相关性来从已知的复合目标对中学习。复合扰动概况和基因敲除概况。在基准集和大型时间分割验证数据集上,与以前的方法(例如连接图)相比,该模型实现了更高的目标推理精度。预测结果的进一步实验验证凸显了 SSGCN 在推断化合物的相互作用靶点或相反地寻找给定感兴趣靶点的新型抑制剂方面的实际用途。











































 京公网安备 11010802027423号
京公网安备 11010802027423号