当前位置:
X-MOL 学术
›
J. Chem. Theory Comput.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Tumuc1: A New Accurate DNA Force Field Consistent with High-Level Quantum Chemistry
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2021-10-18 , DOI: 10.1021/acs.jctc.1c00682 Korbinian Liebl 1 , Martin Zacharias 1
Journal of Chemical Theory and Computation ( IF 5.7 ) Pub Date : 2021-10-18 , DOI: 10.1021/acs.jctc.1c00682 Korbinian Liebl 1 , Martin Zacharias 1
Affiliation
|
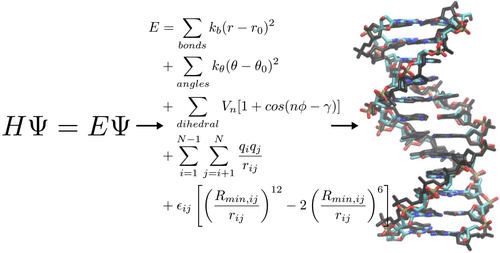
An accurate molecular mechanics force field forms the basis of Molecular Dynamics simulations to obtain a realistic view of the structure and dynamics of biomolecules such as DNA. Although frequently updated to improve agreement with available experimental data, DNA force fields still rely in part on parameters introduced more than 20 years ago. We have developed an entirely new DNA force field, Tumuc1, derived from quantum mechanical calculations to obtain a consistent set of bonded parameters and partial atomic charges. The performance of the force field was extensively tested on a variety of DNA molecules. It excels in accuracy of B-DNA simulations but also performs very well on other types of DNA structures and structure formation processes such as hairpin folding, duplex formation, and dynamics of DNA–protein complexes. It can complement existing force fields in order to provide an increasingly accurate description of the structure and dynamics of DNA during simulation studies.
中文翻译:

Tumuc1:与高水平量子化学一致的新型精确 DNA 力场
精确的分子力学力场构成了分子动力学模拟的基础,以获得生物分子(例如 DNA)的结构和动力学的真实视图。尽管经常更新以提高与可用实验数据的一致性,但 DNA 力场仍然部分依赖于 20 多年前引入的参数。我们开发了一个全新的 DNA 力场 Tumuc1,它源自量子力学计算,以获得一组一致的键合参数和部分原子电荷。力场的性能在各种 DNA 分子上进行了广泛测试。它在 B-DNA 模拟的准确性方面表现出色,但在其他类型的 DNA 结构和结构形成过程(如发夹折叠、双链体形成和 DNA-蛋白质复合物的动力学)上也表现出色。
更新日期:2021-11-09
中文翻译:

Tumuc1:与高水平量子化学一致的新型精确 DNA 力场
精确的分子力学力场构成了分子动力学模拟的基础,以获得生物分子(例如 DNA)的结构和动力学的真实视图。尽管经常更新以提高与可用实验数据的一致性,但 DNA 力场仍然部分依赖于 20 多年前引入的参数。我们开发了一个全新的 DNA 力场 Tumuc1,它源自量子力学计算,以获得一组一致的键合参数和部分原子电荷。力场的性能在各种 DNA 分子上进行了广泛测试。它在 B-DNA 模拟的准确性方面表现出色,但在其他类型的 DNA 结构和结构形成过程(如发夹折叠、双链体形成和 DNA-蛋白质复合物的动力学)上也表现出色。











































 京公网安备 11010802027423号
京公网安备 11010802027423号