当前位置:
X-MOL 学术
›
J. Chem. Inf. Model.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Kinase Inhibitor Scaffold Hopping with Deep Learning Approaches
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2021-09-29 , DOI: 10.1021/acs.jcim.1c00608 Lizhao Hu 1, 2 , Yuyao Yang 2, 3 , Shuangjia Zheng 4 , Jun Xu 1, 2 , Ting Ran 3 , Hongming Chen 3
Journal of Chemical Information and Modeling ( IF 5.6 ) Pub Date : 2021-09-29 , DOI: 10.1021/acs.jcim.1c00608 Lizhao Hu 1, 2 , Yuyao Yang 2, 3 , Shuangjia Zheng 4 , Jun Xu 1, 2 , Ting Ran 3 , Hongming Chen 3
Affiliation
|
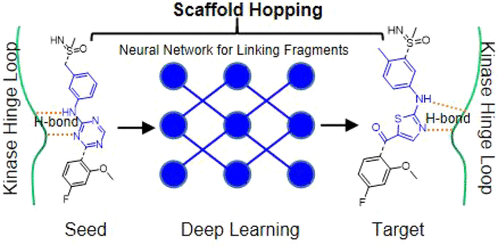
The protein kinase family contains many promising drug targets. Many kinase inhibitors target the ATP-binding pocket, leading to approved drugs in past decades. Scaffold hopping is an effective approach for drug design. The kinase ATP-binding pocket is highly conserved, crossing the whole kinase family. This provides an opportunity to develop a scaffold hopping approach to explore diversified scaffolds among various kinase inhibitors. In this work, we report the SyntaLinker-Hybrid scheme for kinase inhibitor scaffold hopping. With this scheme, we replace molecular fragments bound at the conserved kinase hinge region with deep generative models. Thus, we are able to generate new kinase-inhibitor-like structures hybridizing the privileged fragments against the hinge region. We demonstrate that this scheme allows generation of kinase-inhibitor-like molecules with novel scaffolds, while retaining the binding features of existing kinase inhibitors. This work can be employed in lead identification against kinase targets.
中文翻译:

激酶抑制剂支架使用深度学习方法跳跃
蛋白激酶家族包含许多有前景的药物靶点。许多激酶抑制剂靶向 ATP 结合口袋,导致在过去几十年中获得批准的药物。支架跳跃是一种有效的药物设计方法。激酶 ATP 结合口袋高度保守,跨越整个激酶家族。这为开发支架跳跃方法提供了机会,以探索各种激酶抑制剂之间的多样化支架。在这项工作中,我们报告了激酶抑制剂支架跳跃的 SyntaLinker-Hybrid 方案。通过这个方案,我们用深度生成模型替换了结合在保守激酶铰链区的分子片段。因此,我们能够产生新的激酶抑制剂样结构,将特权片段与铰链区杂交。我们证明该方案允许生成具有新型支架的激酶抑制剂样分子,同时保留现有激酶抑制剂的结合特征。这项工作可用于针对激酶目标的先导识别。
更新日期:2021-10-25
中文翻译:

激酶抑制剂支架使用深度学习方法跳跃
蛋白激酶家族包含许多有前景的药物靶点。许多激酶抑制剂靶向 ATP 结合口袋,导致在过去几十年中获得批准的药物。支架跳跃是一种有效的药物设计方法。激酶 ATP 结合口袋高度保守,跨越整个激酶家族。这为开发支架跳跃方法提供了机会,以探索各种激酶抑制剂之间的多样化支架。在这项工作中,我们报告了激酶抑制剂支架跳跃的 SyntaLinker-Hybrid 方案。通过这个方案,我们用深度生成模型替换了结合在保守激酶铰链区的分子片段。因此,我们能够产生新的激酶抑制剂样结构,将特权片段与铰链区杂交。我们证明该方案允许生成具有新型支架的激酶抑制剂样分子,同时保留现有激酶抑制剂的结合特征。这项工作可用于针对激酶目标的先导识别。











































 京公网安备 11010802027423号
京公网安备 11010802027423号