当前位置:
X-MOL 学术
›
Mol. Syst. Des. Eng.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Probing remdesivir nucleotide analogue insertion to SARS-CoV-2 RNA dependent RNA polymerase in viral replication
Molecular Systems Design & Engineering ( IF 3.2 ) Pub Date : 2021-09-13 , DOI: 10.1039/d1me00088h Moises Ernesto Romero 1 , Chunhong Long 2 , Daniel La Rocco 3 , Anusha Mysore Keerthi 4 , Dajun Xu 5 , Jin Yu 6
Molecular Systems Design & Engineering ( IF 3.2 ) Pub Date : 2021-09-13 , DOI: 10.1039/d1me00088h Moises Ernesto Romero 1 , Chunhong Long 2 , Daniel La Rocco 3 , Anusha Mysore Keerthi 4 , Dajun Xu 5 , Jin Yu 6
Affiliation
|
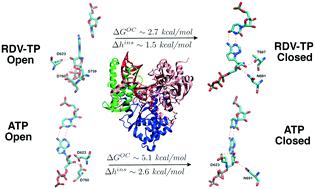
Remdesivir (RDV) prodrug can be metabolized into a triphosphate form nucleotide analogue (RDV-TP) to bind and insert into the active site of viral RNA dependent RNA polymerase (RdRp) to further interfere with viral genome replication. In this work, we computationally studied how RDV-TP binds and inserts to the SARS-CoV-2 RdRp active site, in comparison with natural nucleotide substrate adenosine triphosphate (ATP). To do that, we first constructed atomic structural models of an initial binding complex (active site open) and a substrate insertion complex (active site closed), based on high-resolution cryo-EM structures determined recently for SARS-CoV-2 RdRp or non-structural protein (nsp) 12, in complex with accessory protein factors nsp7 and nsp8. By conducting all-atom molecular dynamics simulation with umbrella sampling strategies on the nucleotide insertion between the open and closed state RdRp complexes, our studies show that RDV-TP can initially bind in a comparatively stabilized state to the viral RdRp active site, as it primarily forms base stacking with the template uracil nucleotide (nt +1), which under freely fluctuations supports a low free energy barrier of the RDV-TP insertion (∼1.5 kcal mol−1). In comparison, the corresponding natural substrate ATP binds initially to the RdRp active site in Watson–Crick base pairing with the template nt, and inserts into the active site with a medium low free energy barrier (∼2.6 kcal mol−1), when the fluctuations of the template nt are well quenched. The simulations also show that the initial base stacking of RDV-TP with the template can be specifically stabilized by motif C-S759, S682 (near motif B) with the base, and motif G-K500 with the template backbone. Although the RDV-TP insertion can be hindered by motif F-R555/R553 interaction with the triphosphate, the ATP insertion seems to be facilitated by such interactions. The inserted RDV-TP and ATP can be further distinguished by specific sugar interaction with motif B-T687 and motif A-D623, respectively.
中文翻译:

在病毒复制中探索瑞德西韦核苷酸类似物插入 SARS-CoV-2 RNA 依赖的 RNA 聚合酶
瑞德西韦 (RDV) 前药可以代谢成三磷酸形式的核苷酸类似物 (RDV-TP),结合并插入病毒 RNA 依赖性 RNA 聚合酶 (RdRp) 的活性位点,进一步干扰病毒基因组复制。在这项工作中,与天然核苷酸底物三磷酸腺苷 (ATP) 相比,我们通过计算研究了 RDV-TP 如何结合并插入 SARS-CoV-2 RdRp 活性位点。为此,我们首先基于最近为 SARS-CoV-2 RdRp 或非结构蛋白 (nsp) 12,与辅助蛋白因子 nsp7 和 nsp8 复合。-1 )。相比之下,相应的天然底物 ATP 最初与 Watson-Crick 碱基配对中的 RdRp 活性位点与模板 nt 结合,并以中等低自由能垒(~2.6 kcal mol -1),当模板nt的波动被很好地抑制时。模拟还表明,RDV-TP 与模板的初始碱基堆叠可以通过带有碱基的基序 C-S759、S682(靠近基序 B)和带有模板骨架的基序 G-K500 来特别稳定。尽管 RDV-TP 插入会受到基序 F-R555/R553 与三磷酸的相互作用的阻碍,但这种相互作用似乎促进了 ATP 的插入。插入的 RDV-TP 和 ATP 可以通过分别与基序 B-T687 和基序 A-D623 的特定糖相互作用进一步区分。
更新日期:2021-09-30
中文翻译:

在病毒复制中探索瑞德西韦核苷酸类似物插入 SARS-CoV-2 RNA 依赖的 RNA 聚合酶
瑞德西韦 (RDV) 前药可以代谢成三磷酸形式的核苷酸类似物 (RDV-TP),结合并插入病毒 RNA 依赖性 RNA 聚合酶 (RdRp) 的活性位点,进一步干扰病毒基因组复制。在这项工作中,与天然核苷酸底物三磷酸腺苷 (ATP) 相比,我们通过计算研究了 RDV-TP 如何结合并插入 SARS-CoV-2 RdRp 活性位点。为此,我们首先基于最近为 SARS-CoV-2 RdRp 或非结构蛋白 (nsp) 12,与辅助蛋白因子 nsp7 和 nsp8 复合。-1 )。相比之下,相应的天然底物 ATP 最初与 Watson-Crick 碱基配对中的 RdRp 活性位点与模板 nt 结合,并以中等低自由能垒(~2.6 kcal mol -1),当模板nt的波动被很好地抑制时。模拟还表明,RDV-TP 与模板的初始碱基堆叠可以通过带有碱基的基序 C-S759、S682(靠近基序 B)和带有模板骨架的基序 G-K500 来特别稳定。尽管 RDV-TP 插入会受到基序 F-R555/R553 与三磷酸的相互作用的阻碍,但这种相互作用似乎促进了 ATP 的插入。插入的 RDV-TP 和 ATP 可以通过分别与基序 B-T687 和基序 A-D623 的特定糖相互作用进一步区分。











































 京公网安备 11010802027423号
京公网安备 11010802027423号