当前位置:
X-MOL 学术
›
Chem. Sci.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Modelling the active SARS-CoV-2 helicase complex as a basis for structure-based inhibitor design
Chemical Science ( IF 7.6 ) Pub Date : 2021-09-06 , DOI: 10.1039/d1sc02775a Dénes Berta 1, 2 , Magd Badaoui 1, 2 , Sam Alexander Martino 1, 2 , Pedro J Buigues 1, 2 , Andrei V Pisliakov 3 , Nadia Elghobashi-Meinhardt 4 , Geoff Wells 5 , Sarah A Harris 6 , Elisa Frezza 7 , Edina Rosta 1, 2
Chemical Science ( IF 7.6 ) Pub Date : 2021-09-06 , DOI: 10.1039/d1sc02775a Dénes Berta 1, 2 , Magd Badaoui 1, 2 , Sam Alexander Martino 1, 2 , Pedro J Buigues 1, 2 , Andrei V Pisliakov 3 , Nadia Elghobashi-Meinhardt 4 , Geoff Wells 5 , Sarah A Harris 6 , Elisa Frezza 7 , Edina Rosta 1, 2
Affiliation
|
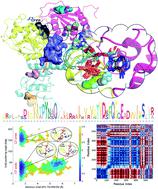
The RNA helicase (non-structural protein 13, NSP13) of SARS-CoV-2 is essential for viral replication, and it is highly conserved among the coronaviridae family, thus a prominent drug target to treat COVID-19. We present here structural models and dynamics of the helicase in complex with its native substrates based on thorough analysis of homologous sequences and existing experimental structures. We performed and analysed microseconds of molecular dynamics (MD) simulations, and our model provides valuable insights to the binding of the ATP and ssRNA at the atomic level. We identify the principal motions characterising the enzyme and highlight the effect of the natural substrates on this dynamics. Furthermore, allosteric binding sites are suggested by our pocket analysis. Our obtained structural and dynamical insights are important for subsequent studies of the catalytic function and for the development of specific inhibitors at our characterised binding pockets for this promising COVID-19 drug target.
中文翻译:

对活性 SARS-CoV-2 解旋酶复合物进行建模,作为基于结构的抑制剂设计的基础
SARS-CoV-2 的 RNA 解旋酶(非结构蛋白 13,NSP13)对于病毒复制至关重要,并且在冠状病毒科中高度保守,因此是治疗 COVID-19 的重要药物靶点。基于对同源序列和现有实验结构的彻底分析,我们在此提出了解旋酶与其天然底物复合物的结构模型和动力学。我们执行并分析了微秒级的分子动力学 (MD) 模拟,我们的模型为原子水平上 ATP 和 ssRNA 的结合提供了宝贵的见解。我们确定了酶的主要运动特征,并强调了天然底物对该动力学的影响。此外,我们的口袋分析表明了变构结合位点。我们获得的结构和动力学见解对于催化功能的后续研究以及针对这一有前景的 COVID-19 药物靶点在我们的特征结合口袋中开发特异性抑制剂非常重要。
更新日期:2021-09-27
中文翻译:

对活性 SARS-CoV-2 解旋酶复合物进行建模,作为基于结构的抑制剂设计的基础
SARS-CoV-2 的 RNA 解旋酶(非结构蛋白 13,NSP13)对于病毒复制至关重要,并且在冠状病毒科中高度保守,因此是治疗 COVID-19 的重要药物靶点。基于对同源序列和现有实验结构的彻底分析,我们在此提出了解旋酶与其天然底物复合物的结构模型和动力学。我们执行并分析了微秒级的分子动力学 (MD) 模拟,我们的模型为原子水平上 ATP 和 ssRNA 的结合提供了宝贵的见解。我们确定了酶的主要运动特征,并强调了天然底物对该动力学的影响。此外,我们的口袋分析表明了变构结合位点。我们获得的结构和动力学见解对于催化功能的后续研究以及针对这一有前景的 COVID-19 药物靶点在我们的特征结合口袋中开发特异性抑制剂非常重要。











































 京公网安备 11010802027423号
京公网安备 11010802027423号