当前位置:
X-MOL 学术
›
Anal. Chem.
›
论文详情
Our official English website, www.x-mol.net, welcomes your feedback! (Note: you will need to create a separate account there.)
High-Resolution Mapping of Amino Acid Residues in DNA–Protein Cross-Links Enabled by Ribonucleotide-Containing DNA
Analytical Chemistry ( IF 7.4 ) Pub Date : 2021-09-24 , DOI: 10.1021/acs.analchem.1c03481 Jin Tang 1 , Wenxin Zhao 1 , Nathan G Hendricks 2 , Linlin Zhao 1, 3
Analytical Chemistry ( IF 7.4 ) Pub Date : 2021-09-24 , DOI: 10.1021/acs.analchem.1c03481 Jin Tang 1 , Wenxin Zhao 1 , Nathan G Hendricks 2 , Linlin Zhao 1, 3
Affiliation
|
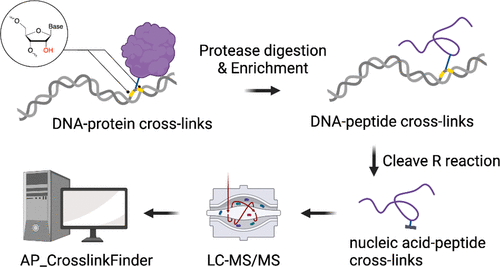
DNA–protein cross-links have broad applications in mapping DNA–protein interactions and provide structural insights into macromolecular structures. However, high-resolution mapping of DNA-interacting amino acid residues with tandem mass spectrometry remains challenging due to difficulties in sample preparation and data analysis. Herein, we developed a method for identifying cross-linking amino residues in DNA–protein cross-links at single amino acid resolution. We leveraged the alkaline lability of ribonucleotides and designed ribonucleotide-containing DNA to produce structurally defined nucleic acid–peptide cross-links under our optimized ribonucleotide cleavage conditions. The structurally defined oligonucleotide–peptide heteroconjugates improved ionization, reduced the database search space, and facilitated the identification of cross-linking residues in peptides. We applied the workflow to identifying abasic (AP) site-interacting residues in human mitochondrial transcription factor A (TFAM)-DNA cross-links. With sub-nmol sample input, we obtained high-quality fragmentation spectra for nucleic acid–peptide cross-links and identified 14 cross-linked lysine residues with the home-built AP_CrosslinkFinder program. Semi-quantification based on integrated peak areas revealed that K186 of TFAM is the major cross-linking residue, consistent with K186 being the closest (to the AP modification) lysine residue in solved TFAM:DNA crystal structures. Additional cross-linking lysine residues (K69, K76, K136, K154) support the dynamic characteristics of TFAM:DNA complexes. Overall, our combined workflow using ribonucleotide as a chemically cleavable DNA modification together with optimized sample preparation and data analysis offers a simple yet powerful approach for mapping cross-linking sites in DNA–protein cross-links. The method is amendable to other chemical or photo-cross-linking systems and can be extended to complex biological samples.
中文翻译:

由含有核糖核苷酸的 DNA 实现的 DNA-蛋白质交联中氨基酸残基的高分辨率图谱
DNA-蛋白质交联在绘制 DNA-蛋白质相互作用图和提供对大分子结构的结构洞察方面具有广泛的应用。然而,由于样品制备和数据分析的困难,使用串联质谱法对 DNA 相互作用的氨基酸残基进行高分辨率定位仍然具有挑战性。在此,我们开发了一种以单氨基酸分辨率识别 DNA-蛋白质交联中交联氨基残基的方法。我们利用核糖核苷酸的碱性不稳定性并设计了含有核糖核苷酸的 DNA,以在我们优化的核糖核苷酸切割条件下产生结构明确的核酸-肽交联。结构确定的寡核苷酸-肽杂合物改善了离子化,减少了数据库搜索空间,并促进肽中交联残基的鉴定。我们将工作流程应用于识别人线粒体转录因子 A (TFAM)-DNA 交联中的无碱基 (AP) 位点相互作用残基。通过亚 nmol 样品输入,我们获得了核酸-肽交联的高质量碎片光谱,并使用自制的 AP_CrosslinkFinder 程序识别了 14 个交联的赖氨酸残基。基于积分峰面积的半定量显示 TFAM 的 K186 是主要的交联残基,这与 K186 是已解决的 TFAM:DNA 晶体结构中最接近(最接近 AP 修饰)的赖氨酸残基一致。额外的交联赖氨酸残基(K69、K76、K136、K154)支持 TFAM:DNA 复合物的动态特性。全面的,我们使用核糖核苷酸作为可化学切割的 DNA 修饰的组合工作流程以及优化的样品制备和数据分析为绘制 DNA-蛋白质交联中的交联位点提供了一种简单而强大的方法。该方法适用于其他化学或光交联系统,并可扩展到复杂的生物样品。
更新日期:2021-10-06
中文翻译:

由含有核糖核苷酸的 DNA 实现的 DNA-蛋白质交联中氨基酸残基的高分辨率图谱
DNA-蛋白质交联在绘制 DNA-蛋白质相互作用图和提供对大分子结构的结构洞察方面具有广泛的应用。然而,由于样品制备和数据分析的困难,使用串联质谱法对 DNA 相互作用的氨基酸残基进行高分辨率定位仍然具有挑战性。在此,我们开发了一种以单氨基酸分辨率识别 DNA-蛋白质交联中交联氨基残基的方法。我们利用核糖核苷酸的碱性不稳定性并设计了含有核糖核苷酸的 DNA,以在我们优化的核糖核苷酸切割条件下产生结构明确的核酸-肽交联。结构确定的寡核苷酸-肽杂合物改善了离子化,减少了数据库搜索空间,并促进肽中交联残基的鉴定。我们将工作流程应用于识别人线粒体转录因子 A (TFAM)-DNA 交联中的无碱基 (AP) 位点相互作用残基。通过亚 nmol 样品输入,我们获得了核酸-肽交联的高质量碎片光谱,并使用自制的 AP_CrosslinkFinder 程序识别了 14 个交联的赖氨酸残基。基于积分峰面积的半定量显示 TFAM 的 K186 是主要的交联残基,这与 K186 是已解决的 TFAM:DNA 晶体结构中最接近(最接近 AP 修饰)的赖氨酸残基一致。额外的交联赖氨酸残基(K69、K76、K136、K154)支持 TFAM:DNA 复合物的动态特性。全面的,我们使用核糖核苷酸作为可化学切割的 DNA 修饰的组合工作流程以及优化的样品制备和数据分析为绘制 DNA-蛋白质交联中的交联位点提供了一种简单而强大的方法。该方法适用于其他化学或光交联系统,并可扩展到复杂的生物样品。



























 京公网安备 11010802027423号
京公网安备 11010802027423号