Letters in Drug Design & Discovery ( IF 1.2 ) Pub Date : 2021-06-30 , DOI: 10.2174/1570180817999201211192211 Carlos Andrés Rodríguez-Salazar 1 , Delia Piedad Recalde-Reyes 1 , Jhon Carlos Castaño-Osorio 2
|
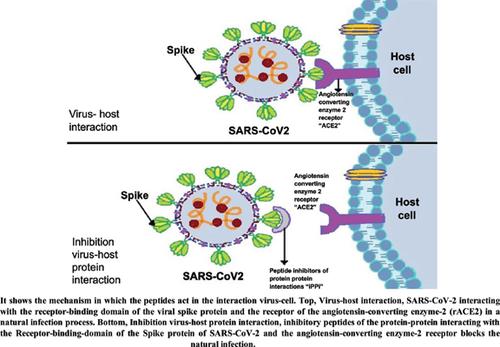
Background: The recent outbreak caused by SARS-CoV-2, known as COVID-19, has been cataloged as a global catastrophe due to the growing number of infected cases and deaths since November 2019; this infectious, contagious disease, to date, does not have a vaccine or specific treatment available, which is why the number of cases continues to increase. SARS-CoV-2 infects humans as a result of the interaction between the receptor-binding domain of the viral spike protein and the receptor of the angiotensin-converting enzyme-2 (rACE2), located predominantly in the alveolar cells.
Objective: This study aims to identify inhibitory peptides of the protein-protein interaction between the receptor-binding-domain of the spike protein of SARS-CoV-2 and the angiotensin-converting enzyme-2 receptor through computational tools.
Methods: Through the Research Collaboratory for Structural Bioinformatics protein database, crystals were selected and interaction models were carried out between the viral protein and the ACE2; thereafter, the study designed inhibitory peptides of the interaction through the Rosetta web server, validated their interaction through ClusPro and, finally, determined the theoretical physicochemical and cytotoxic properties.
Results: A protein complex was generated and modeled through ClusPro; the balanced model was selected with the lowest binding energy. From the protein interactions of each of the crystals and from the model, eight peptides of 20 residues were obtained. The theoretical evaluation showed non-toxic peptides, six soluble in water, and two insoluble.
Conclusion: We found eight peptides that interacted with the receptor-binding-domain of the spike protein of SARS-CoV-2, which could avoid contact with the cell receptor and generate interference in the infection process.
中文翻译:

SARS-CoV-2 S1蛋白RBD结构域与血管紧张素转换酶2(ACE2)受体相互作用抑制肽的设计
背景:最近由 SARS-CoV-2 引起的爆发,即 COVID-19,由于自 2019 年 11 月以来感染病例和死亡人数不断增加,已被列为全球灾难;迄今为止,这种传染性传染病没有可用的疫苗或特定治疗方法,这就是病例数不断增加的原因。由于病毒刺突蛋白的受体结合域与主要位于肺泡细胞中的血管紧张素转换酶-2 (rACE2) 受体之间的相互作用,SARS-CoV-2 感染了人类。
目的:本研究旨在通过计算工具鉴定 SARS-CoV-2 刺突蛋白的受体结合结构域与血管紧张素转换酶 2 受体之间蛋白质-蛋白质相互作用的抑制肽。
方法:通过结构生物信息学蛋白质数据库研究合作实验室,选择晶体并建立病毒蛋白与ACE2之间的相互作用模型;此后,该研究通过 Rosetta 网络服务器设计了相互作用的抑制肽,通过 ClusPro 验证了它们的相互作用,最后确定了理论物理化学和细胞毒性特性。
结果:通过ClusPro生成并建模了蛋白质复合物;选择结合能最低的平衡模型。从每个晶体的蛋白质相互作用和模型中,获得了 8 个 20 个残基的肽。理论评估表明肽是无毒的,六种可溶于水,两种不溶于水。
结论:我们发现8个肽段与SARS-CoV-2刺突蛋白的受体结合域相互作用,可以避免与细胞受体接触,对感染过程产生干扰。










































 京公网安备 11010802027423号
京公网安备 11010802027423号