Water Research ( IF 11.4 ) Pub Date : 2021-09-23 , DOI: 10.1016/j.watres.2021.117681 Tyson E Graber 1 , Élisabeth Mercier 2 , Kamya Bhatnagar 2 , Meghan Fuzzen 3 , Patrick M D'Aoust 2 , Huy-Dung Hoang 4 , Xin Tian 2 , Syeda Tasneem Towhid 2 , Julio Plaza-Diaz 1 , Walaa Eid 1 , Tommy Alain 4 , Ainslie Butler 5 , Lawrence Goodridge 6 , Mark Servos 3 , Robert Delatolla 2
|
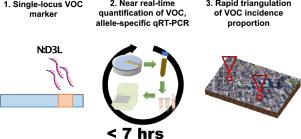
The coronavirus disease 2019 (COVID-19) pandemic caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has claimed millions of lives to date. Antigenic drift has resulted in viral variants with putatively greater transmissibility, virulence, or both. Early and near real-time detection of these variants of concern (VOC) and the ability to accurately follow their incidence and prevalence in communities is wanting. Wastewater-based epidemiology (WBE), which uses nucleic acid amplification tests to detect viral fragments, is a reliable proxy of COVID-19 incidence and prevalence, and thus offers the potential to monitor VOC viral load in a given population. Here, we describe and validate a primer extension PCR strategy targeting a signature mutation in the N gene of SARS-CoV-2. This allows quantification of B.1.1.7 versus non-B.1.1.7 allele frequency in wastewater without the need to employ quantitative RT-PCR standard curves. We show that the wastewater B.1.1.7 profile correlates with its clinical counterpart and benefits from a near real-time and facile data collection and reporting pipeline. This assay can be quickly implemented within a current SARS-CoV-2 WBE framework with minimal cost; allowing early and contemporaneous estimates of B.1.1.7 community transmission prior to, or in lieu of, clinical screening and identification. Our study demonstrates that this strategy can provide public health units with an additional and much needed tool to rapidly triangulate VOC incidence/prevalence with high sensitivity and lineage specificity.
中文翻译:

使用等位基因特异性引物延伸 PCR 策略近实时测定 B.1.1.7 与废水中总 SARS-CoV-2 病毒载量的比例
迄今为止,由严重急性呼吸系统综合症冠状病毒 2 (SARS-CoV-2) 引起的 2019 年冠状病毒病 (COVID-19) 大流行已夺去数百万人的生命。抗原漂移已导致病毒变体具有更高的传播能力和/或毒力。需要对这些关注的变体 (VOC) 进行早期和近乎实时的检测,并能够准确跟踪它们在社区中的发生率和流行程度。基于废水的流行病学 (WBE) 使用核酸扩增测试来检测病毒片段,是 COVID-19 发病率和流行率的可靠代表,因此有可能监测特定人群中的 VOC 病毒载量。在这里,我们描述并验证了针对 SARS-CoV-2 N 基因特征突变的引物延伸 PCR 策略。这允许量化 B.1.1.7 与非 B.1.1。废水中的 7 等位基因频率,无需使用定量 RT-PCR 标准曲线。我们表明,废水 B.1.1.7 概况与其临床对应物相关,并受益于近乎实时和简便的数据收集和报告管道。该检测可以在当前的 SARS-CoV-2 WBE 框架内以最低成本快速实施;B.1.1.7 允许在临床筛查和鉴定之前或代替临床筛查和鉴定对社区传播进行早期和同期估计。我们的研究表明,该策略可以为公共卫生部门提供一种额外且急需的工具,以高灵敏度和谱系特异性快速三角测量 VOC 发生率/流行率。7 profile 与其临床对应物相关联,并受益于近乎实时和简便的数据收集和报告管道。该检测可以在当前的 SARS-CoV-2 WBE 框架内以最低成本快速实施;B.1.1.7 允许在临床筛查和鉴定之前或代替临床筛查和鉴定对社区传播进行早期和同期估计。我们的研究表明,该策略可以为公共卫生部门提供一种额外且急需的工具,以高灵敏度和谱系特异性快速三角测量 VOC 发生率/流行率。7 profile 与其临床对应物相关联,并受益于近乎实时和简便的数据收集和报告管道。该检测可以在当前的 SARS-CoV-2 WBE 框架内以最低成本快速实施;B.1.1.7 允许在临床筛查和鉴定之前或代替临床筛查和鉴定对社区传播进行早期和同期估计。我们的研究表明,该策略可以为公共卫生部门提供一种额外且急需的工具,以高灵敏度和谱系特异性快速三角测量 VOC 发生率/流行率。B.1.1.7 允许在临床筛查和鉴定之前或代替临床筛查和鉴定对社区传播进行早期和同期估计。我们的研究表明,该策略可以为公共卫生部门提供一种额外且急需的工具,以高灵敏度和谱系特异性快速三角测量 VOC 发生率/流行率。B.1.1.7 允许在临床筛查和鉴定之前或代替临床筛查和鉴定对社区传播进行早期和同期估计。我们的研究表明,该策略可以为公共卫生部门提供一种额外且急需的工具,以高灵敏度和谱系特异性快速三角测量 VOC 发生率/流行率。











































 京公网安备 11010802027423号
京公网安备 11010802027423号