Analytica Chimica Acta ( IF 5.7 ) Pub Date : 2021-09-20 , DOI: 10.1016/j.aca.2021.339073 Peter B Skou 1 , Ensie Hosseini 2 , Jahan B Ghasemi 2 , Age K Smilde 3 , Carl Emil Eskildsen 4
|
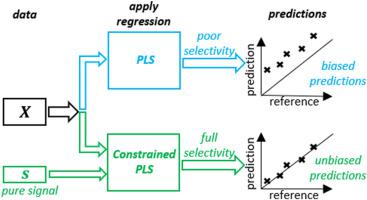
In analytical chemistry spectroscopy is attractive for high-throughput quantification, which often relies on inverse regression, like partial least squares regression. Due to a multivariate nature of spectroscopic measurements an analyte can be quantified in presence of interferences. However, if the model is not fully selective against interferences, analyte predictions may be biased. The degree of model selectivity against an interferent is defined by the inner relation between the regression vector and the pure interfering signal. If the regression vector is orthogonal to the signal, this inner relation equals zero and the model is fully selective. The degree of model selectivity largely depends on calibration data quality. Strong correlations may deteriorate calibration data resulting in poorly selective models. We show this using a fructose-maltose model system. Furthermore, we modify the NIPALS algorithm to improve model selectivity when calibration data are deteriorated. This modification is done by incorporating a projection matrix into the algorithm, which constrains regression vector estimation to the null-space of known interfering signals. This way known interfering signals are handled, while unknown signals are accounted for by latent variables. We test the modified algorithm and compare it to the conventional NIPALS algorithm using both simulated and industrial process data. The industrial process data consist of mid-infrared measurements obtained on mixtures of beta-lactoglobulin (analyte of interest), and alpha-lactalbumin and caseinoglycomacropeptide (interfering species). The root mean squared error of beta-lactoglobulin (% w/w) predictions of a test set was 0.92 and 0.33 when applying the conventional and the modified NIPALS algorithm, respectively. Our modification of the algorithm returns simpler models with improved selectivity and analyte predictions. This paper shows how known interfering signals may be utilized in a direct fashion, while benefitting from a latent variable approach. The modified algorithm can be viewed as a fusion between ordinary least squares regression and partial least squares regression and may be very useful when knowledge of some (but not all) interfering species is available.
中文翻译:

正交约束逆回归以提高模型选择性和振动光谱测量的分析物预测
在分析化学中,光谱对高通量定量很有吸引力,这通常依赖于逆回归,如偏最小二乘回归。由于光谱测量的多变量性质,可以在存在干扰的情况下对分析物进行量化。但是,如果模型对干扰没有完全选择性,分析物预测可能会出现偏差。模型对干扰物的选择性程度由回归向量和纯干扰信号之间的内在关系定义。如果回归向量与信号正交,则该内部关系为零,并且模型是完全选择性的。模型选择性的程度在很大程度上取决于校准数据的质量。强相关性可能会使校准数据变差,从而导致模型选择不佳。我们使用果糖-麦芽糖模型系统展示了这一点。此外,我们修改了 NIPALS 算法以在校准数据恶化时提高模型选择性。这种修改是通过将投影矩阵合并到算法中来完成的,该矩阵将回归向量估计限制为空值-已知干扰信号的空间。以这种方式处理已知干扰信号,而未知信号由潜在变量解释。我们测试修改后的算法,并使用模拟和工业过程数据将其与传统的 NIPALS 算法进行比较。工业过程数据包括对 β-乳球蛋白(目标分析物)、α-乳清蛋白和酪蛋白糖巨肽(干扰物质)的混合物进行的中红外测量。当应用传统和改进的 NIPALS 算法时,测试集的 β-乳球蛋白 (% w/w) 预测的均方根误差分别为 0.92 和 0.33。我们对算法的修改返回了更简单的模型,具有改进的选择性和分析物预测。本文展示了如何以直接方式利用已知干扰信号,同时受益于潜在变量方法。修改后的算法可以被视为普通最小二乘回归和偏最小二乘回归之间的融合,并且在某些(但不是全部)干扰物种的知识可用时可能非常有用。











































 京公网安备 11010802027423号
京公网安备 11010802027423号