当前位置:
X-MOL 学术
›
Comput. Struct. Biotechnol. J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Integrative structural biology of the penicillin-binding protein-1 from Staphylococcus aureus, an essential component of the divisome machinery
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-09-17 , DOI: 10.1016/j.csbj.2021.09.018 Siseth Martínez-Caballero 1 , Kiran V Mahasenan 2 , Choon Kim 2 , Rafael Molina 1 , Rhona Feltzer 2 , Mijoon Lee 2 , Renee Bouley 2 , Dusan Hesek 2 , Jed F Fisher 2 , Inés G Muñoz 3 , Mayland Chang 2 , Shahriar Mobashery 2 , Juan A Hermoso 1
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-09-17 , DOI: 10.1016/j.csbj.2021.09.018 Siseth Martínez-Caballero 1 , Kiran V Mahasenan 2 , Choon Kim 2 , Rafael Molina 1 , Rhona Feltzer 2 , Mijoon Lee 2 , Renee Bouley 2 , Dusan Hesek 2 , Jed F Fisher 2 , Inés G Muñoz 3 , Mayland Chang 2 , Shahriar Mobashery 2 , Juan A Hermoso 1
Affiliation
|
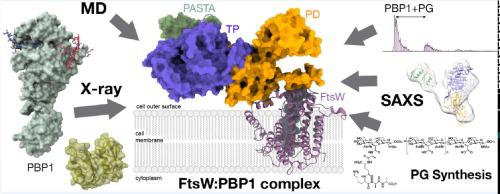
The penicillin-binding proteins are the enzyme catalysts of the critical transpeptidation crosslinking polymerization reaction of bacterial peptidoglycan synthesis and the molecular targets of the penicillin antibiotics. Here, we report a combined crystallographic, small-angle X-ray scattering (SAXS) in-solution structure, computational and biophysical analysis ofPBP1 ofPBP1), providing mechanistic clues about its function and regulation during cell division. The structure reveals the pedestal domain, the transpeptidase domain, and most of the linker connecting to the “penicillin-binding protein and serine/threonine kinase associated” (PASTA) domains, but not its two PASTA domains, despite their presence in the construct. To address this absence, the structure of the PASTA domainswas determined at 1.5 Å resolution. Extensive molecular-dynamics simulations interpret the PASTA domains ofPBP1 as conformationally mobile and separated from the transpeptidasedomain.This conclusion was confirmed by SAXS experiments on the full-length protein in solution.A series of crystallographic complexes with β-lactam antibiotics (as inhibitors) and penta-Gly (as a substrate mimetic) allowed the molecular characterization of both inhibition by antibiotics and binding for the donor and acceptor peptidoglycan strands. Mass-spectrometry experiments with synthetic peptidoglycan fragments revealed binding by PASTA domains in coordination with the remaining domains. The observed mobility of the PASTA domain inPBP1 could play a crucial role forinteraction with its glycosyltransferase partner in the membrane or with other components of the divisome machinery, as well as for coordination of transpeptidation and polymerization processes in the bacterial divisome.
中文翻译:

金黄色葡萄球菌青霉素结合蛋白 1 的整合结构生物学,它是分裂体机制的重要组成部分
青霉素结合蛋白是细菌肽聚糖合成的关键转肽交联聚合反应的酶催化剂,也是青霉素抗生素的分子靶标。在这里,我们报告了 PBP1 (PBP1) 的结合晶体学、小角 X 射线散射 (SAXS) 溶液结构、计算和生物物理分析,提供了有关其在细胞分裂过程中的功能和调节的机制线索。该结构揭示了基座结构域、转肽酶结构域以及连接到“青霉素结合蛋白和丝氨酸/苏氨酸激酶相关”(PASTA) 结构域的大部分接头,但没有显示其两个 PASTA 结构域,尽管它们存在于构建体中。为了解决这一问题,以 1.5 Å 分辨率确定了 PASTA 结构域的结构。广泛的分子动力学模拟将 PBP1 的 PASTA 结构域解释为构象可移动并与转肽酶结构域分离。这一结论通过对溶液中全长蛋白的 SAXS 实验得到了证实。一系列与 β-内酰胺抗生素(作为抑制剂)和五甘氨酸(作为底物模拟物)可以对抗生素的抑制作用以及供体和受体肽聚糖链的结合进行分子表征。合成肽聚糖片段的质谱实验揭示了 PASTA 结构域与其余结构域的结合。观察到的 PBP1 中 PASTA 结构域的移动性对于与其在膜中的糖基转移酶伴侣或与分裂体机器的其他组件的相互作用以及细菌分裂体中转肽和聚合过程的协调发挥着至关重要的作用。
更新日期:2021-09-17
中文翻译:

金黄色葡萄球菌青霉素结合蛋白 1 的整合结构生物学,它是分裂体机制的重要组成部分
青霉素结合蛋白是细菌肽聚糖合成的关键转肽交联聚合反应的酶催化剂,也是青霉素抗生素的分子靶标。在这里,我们报告了 PBP1 (PBP1) 的结合晶体学、小角 X 射线散射 (SAXS) 溶液结构、计算和生物物理分析,提供了有关其在细胞分裂过程中的功能和调节的机制线索。该结构揭示了基座结构域、转肽酶结构域以及连接到“青霉素结合蛋白和丝氨酸/苏氨酸激酶相关”(PASTA) 结构域的大部分接头,但没有显示其两个 PASTA 结构域,尽管它们存在于构建体中。为了解决这一问题,以 1.5 Å 分辨率确定了 PASTA 结构域的结构。广泛的分子动力学模拟将 PBP1 的 PASTA 结构域解释为构象可移动并与转肽酶结构域分离。这一结论通过对溶液中全长蛋白的 SAXS 实验得到了证实。一系列与 β-内酰胺抗生素(作为抑制剂)和五甘氨酸(作为底物模拟物)可以对抗生素的抑制作用以及供体和受体肽聚糖链的结合进行分子表征。合成肽聚糖片段的质谱实验揭示了 PASTA 结构域与其余结构域的结合。观察到的 PBP1 中 PASTA 结构域的移动性对于与其在膜中的糖基转移酶伴侣或与分裂体机器的其他组件的相互作用以及细菌分裂体中转肽和聚合过程的协调发挥着至关重要的作用。










































 京公网安备 11010802027423号
京公网安备 11010802027423号