当前位置:
X-MOL 学术
›
Comput. Struct. Biotechnol. J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Structure-guided machine learning prediction of drug resistance mutations in Abelson 1 kinase
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-09-16 , DOI: 10.1016/j.csbj.2021.09.016 Yunzhuo Zhou 1, 2 , Stephanie Portelli 1, 2 , Megan Pat 1, 2 , Carlos H M Rodrigues 1, 2, 3 , Thanh-Binh Nguyen 1, 2, 3, 4 , Douglas E V Pires 1, 2, 3, 5 , David B Ascher 1, 2, 3, 4, 6
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-09-16 , DOI: 10.1016/j.csbj.2021.09.016 Yunzhuo Zhou 1, 2 , Stephanie Portelli 1, 2 , Megan Pat 1, 2 , Carlos H M Rodrigues 1, 2, 3 , Thanh-Binh Nguyen 1, 2, 3, 4 , Douglas E V Pires 1, 2, 3, 5 , David B Ascher 1, 2, 3, 4, 6
Affiliation
|
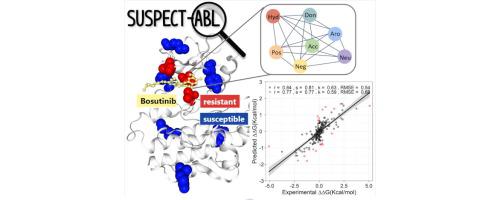
Kinases play crucial roles in cellular signalling and biological processes with their dysregulation associated with diseases, including cancers. Kinase inhibitors, most notably those targeting ABeLson 1 (ABL1) kinase in chronic myeloid leukemia, have had a significant impact on cancer survival, yet emergence of resistance mutations can reduce their effectiveness, leading to therapeutic failure. Limited effort, however, has been devoted to developing tools to accurately identify ABL1 resistance mutations, as well as providing insights into their molecular mechanisms. Here we investigated the structural basis of ABL1 mutations modulating binding affinity of eight FDA-approved drugs. We found mutations impair affinity of type I and type II inhibitors differently and used this insight to developed a novel web-based diagnostic tool, SUSPECT-ABL, to pre-emptively predict resistance profiles and binding free-energy changes (ΔΔ) of all possible ABL1 mutations against inhibitors with different binding modes. Resistance mutations in ABL1 were successfully identified, achieving a Matthew’s Correlation Coefficient of up to 0.73 and the resulting change in ligand binding affinity with a Pearson’s correlation of up to 0.77, with performances consistent across non-redundant blind tests. Through an saturation mutagenesis, our tool has identified possibly emerging resistance mutations, which offers opportunities for experimental validation. We believe SUSPECT-ABL will be an important tool not just for improving precision medicine efforts, but for facilitating the development of next-generation inhibitors that are less prone to resistance. We have made our tool freely available at .
中文翻译:

Abelson 1 激酶耐药突变的结构引导机器学习预测
激酶在细胞信号传导和生物过程中发挥着至关重要的作用,其失调与疾病(包括癌症)相关。激酶抑制剂,尤其是针对慢性粒细胞白血病中的 ABeLson 1 (ABL1) 激酶的抑制剂,对癌症生存具有重大影响,但耐药突变的出现会降低其有效性,导致治疗失败。然而,在开发准确识别 ABL1 抗性突变的工具以及提供对其分子机制的见解方面的努力有限。在这里,我们研究了 ABL1 突变调节八种 FDA 批准药物的结合亲和力的结构基础。我们发现突变以不同的方式损害 I 型和 II 型抑制剂的亲和力,并利用这一见解开发了一种新型的基于网络的诊断工具 SUSPECT-ABL,以预先预测耐药性概况和所有可能的结合自由能变化 (ΔΔ) ABL1 突变针对不同结合模式的抑制剂。成功鉴定了 ABL1 中的耐药突变,马修相关系数高达 0.73,配体结合亲和力发生变化,皮尔逊相关系数高达 0.77,在非冗余盲测中表现一致。通过饱和诱变,我们的工具已经识别出可能出现的抗性突变,这为实验验证提供了机会。我们相信 SUSPECT-ABL 将成为一个重要工具,不仅可以改善精准医疗工作,而且可以促进不易产生耐药性的下一代抑制剂的开发。我们已在 上免费提供我们的工具。
更新日期:2021-09-16
中文翻译:

Abelson 1 激酶耐药突变的结构引导机器学习预测
激酶在细胞信号传导和生物过程中发挥着至关重要的作用,其失调与疾病(包括癌症)相关。激酶抑制剂,尤其是针对慢性粒细胞白血病中的 ABeLson 1 (ABL1) 激酶的抑制剂,对癌症生存具有重大影响,但耐药突变的出现会降低其有效性,导致治疗失败。然而,在开发准确识别 ABL1 抗性突变的工具以及提供对其分子机制的见解方面的努力有限。在这里,我们研究了 ABL1 突变调节八种 FDA 批准药物的结合亲和力的结构基础。我们发现突变以不同的方式损害 I 型和 II 型抑制剂的亲和力,并利用这一见解开发了一种新型的基于网络的诊断工具 SUSPECT-ABL,以预先预测耐药性概况和所有可能的结合自由能变化 (ΔΔ) ABL1 突变针对不同结合模式的抑制剂。成功鉴定了 ABL1 中的耐药突变,马修相关系数高达 0.73,配体结合亲和力发生变化,皮尔逊相关系数高达 0.77,在非冗余盲测中表现一致。通过饱和诱变,我们的工具已经识别出可能出现的抗性突变,这为实验验证提供了机会。我们相信 SUSPECT-ABL 将成为一个重要工具,不仅可以改善精准医疗工作,而且可以促进不易产生耐药性的下一代抑制剂的开发。我们已在 上免费提供我们的工具。











































 京公网安备 11010802027423号
京公网安备 11010802027423号