当前位置:
X-MOL 学术
›
Comput. Struct. Biotechnol. J.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Reconstructing Boolean network ensembles from single-cell data for unraveling dynamics in the aging of human hematopoietic stem cells
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-09-15 , DOI: 10.1016/j.csbj.2021.09.012 Julian D Schwab 1 , Nensi Ikonomi 1 , Silke D Werle 1 , Felix M Weidner 1 , Hartmut Geiger 2 , Hans A Kestler 1
Computational and Structural Biotechnology Journal ( IF 4.4 ) Pub Date : 2021-09-15 , DOI: 10.1016/j.csbj.2021.09.012 Julian D Schwab 1 , Nensi Ikonomi 1 , Silke D Werle 1 , Felix M Weidner 1 , Hartmut Geiger 2 , Hans A Kestler 1
Affiliation
|
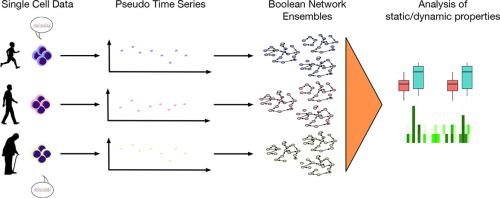
Regulatory dependencies in molecular networks are the basis of dynamic behaviors affecting the phenotypical landscape. With the advance of high throughput technologies, the detail of omics data has arrived at the single-cell level. Nevertheless, new strategies are required to reconstruct regulatory networks based on populations of single-cell data. Here, we present a new approach to generate populations of gene regulatory networks from single-cell RNA-sequencing (scRNA-seq) data. Our approach exploits the heterogeneity of single-cell populations to generate pseudo-timepoints. This allows for the first time to uncouple network reconstruction from a direct dependency on time series measurements. The generated time series are then fed to a combined reconstruction algorithm. The latter allows a fast and efficient reconstruction of ensembles of gene regulatory networks. Since this approach does not require knowledge on time-related trajectories, it allows us to model heterogeneous processes such as aging. Applying the approach to the aging-associated NF-B signaling pathway-based scRNA-seq data of human hematopoietic stem cells (HSCs), we were able to reconstruct eight ensembles, and evaluate their dynamic behavior. Moreover, we propose a strategy to evaluate the resulting attractor patterns. Interaction graph-based features and dynamic investigations of our model ensembles provide a new perspective on the heterogeneity and mechanisms related to human HSCs aging.
中文翻译:

从单细胞数据重建布尔网络集成,以揭示人类造血干细胞衰老的动力学
分子网络中的调控依赖性是影响表型景观的动态行为的基础。随着高通量技术的进步,组学数据的细节已达到单细胞水平。然而,需要新的策略来重建基于单细胞数据群体的调控网络。在这里,我们提出了一种从单细胞 RNA 测序 (scRNA-seq) 数据生成基因调控网络群体的新方法。我们的方法利用单细胞群体的异质性来生成伪时间点。这首次将网络重建与对时间序列测量的直接依赖分开。然后将生成的时间序列输入组合重建算法。后者允许快速有效地重建基因调控网络的整体。由于这种方法不需要有关时间相关轨迹的知识,因此它允许我们对衰老等异构过程进行建模。将该方法应用于基于衰老相关 NF-B 信号通路的人类造血干细胞 (HSC) 的 scRNA-seq 数据,我们能够重建八个集合,并评估它们的动态行为。此外,我们提出了一种评估所产生的吸引子模式的策略。基于交互图的特征和模型集合的动态研究为人类 HSC 衰老相关的异质性和机制提供了新的视角。
更新日期:2021-09-15
中文翻译:

从单细胞数据重建布尔网络集成,以揭示人类造血干细胞衰老的动力学
分子网络中的调控依赖性是影响表型景观的动态行为的基础。随着高通量技术的进步,组学数据的细节已达到单细胞水平。然而,需要新的策略来重建基于单细胞数据群体的调控网络。在这里,我们提出了一种从单细胞 RNA 测序 (scRNA-seq) 数据生成基因调控网络群体的新方法。我们的方法利用单细胞群体的异质性来生成伪时间点。这首次将网络重建与对时间序列测量的直接依赖分开。然后将生成的时间序列输入组合重建算法。后者允许快速有效地重建基因调控网络的整体。由于这种方法不需要有关时间相关轨迹的知识,因此它允许我们对衰老等异构过程进行建模。将该方法应用于基于衰老相关 NF-B 信号通路的人类造血干细胞 (HSC) 的 scRNA-seq 数据,我们能够重建八个集合,并评估它们的动态行为。此外,我们提出了一种评估所产生的吸引子模式的策略。基于交互图的特征和模型集合的动态研究为人类 HSC 衰老相关的异质性和机制提供了新的视角。











































 京公网安备 11010802027423号
京公网安备 11010802027423号