Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Quantifying the force in flow-cell based single-molecule stretching experiments
Nanoscale ( IF 5.8 ) Pub Date : 2021-09-15 , DOI: 10.1039/d1nr04748e Jialun Liang 1, 2 , Jiaxi Li 1, 2 , Zhensheng Zhong 1, 2 , Thitima Rujiralai 3 , Jie Ma 1, 2
Nanoscale ( IF 5.8 ) Pub Date : 2021-09-15 , DOI: 10.1039/d1nr04748e Jialun Liang 1, 2 , Jiaxi Li 1, 2 , Zhensheng Zhong 1, 2 , Thitima Rujiralai 3 , Jie Ma 1, 2
Affiliation
|
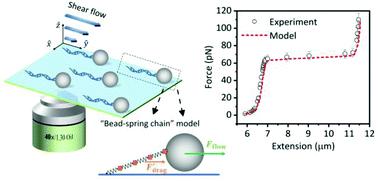
The flow-cell based single-molecule manipulation technique has found many applications in the study of DNA mechanics and protein–DNA interactions. However, the force in these experiments has not been fully characterized and is usually limited to a moderate force regime (<25 pN). In this work, using the “tethered-bead” assay, the hydrodynamic drag of DNA has been quantitatively evaluated based on a “bead-spring chain” model. The force derived from the Brownian motion of the bead thus contains both contributions from this equivalent hydrodynamic drag of DNA and the pulling force from the tethered bead. Next, using flow-cell based DNA pulling experiments, the linear relationship between the flow rate and total hydrodynamic force on the bead-DNA system has been demonstrated to be valid over a wide force range (0–110 pN). Consequently, the force can be directly converted from the flow rate by a linear factor that can be calibrated either by the bead's Brownian motion at low flow rates or using DNA overstretching transition. Furthermore, the hydrodynamic force and torque due to the shear flow on the bead as well as the equivalent stretching force on DNA are calculated based on theoretical models with the hydrodynamic drag on DNA also considered. The calculated force–extension curves show a good agreement with the measured ones. These results offer important insights into the force in flow-cell based single-molecule stretching experiments and provide a foundation for establishing flow-cells as a simple, low-cost, yet flexible and precise tool for single-molecule force measurements over a wide force range.
中文翻译:

量化基于流动池的单分子拉伸实验中的力
基于流动池的单分子操作技术在 DNA 力学和蛋白质-DNA 相互作用的研究中有许多应用。然而,这些实验中的力尚未完全表征,通常仅限于中等力制度 (<25 pN)。在这项工作中,使用“系留珠”测定法,基于“珠-弹簧链”模型定量评估了 DNA 的流体动力学阻力。因此,源自珠子布朗运动的力既包含来自 DNA 的等效流体动力学阻力的贡献,也包含来自系留珠子的拉力。接下来,使用基于流动池的 DNA 拉动实验,流量和珠子 DNA 系统上的总流体动力之间的线性关系已被证明在很宽的力范围 (0–110 pN) 内是有效的。最后,力可以通过线性因子直接从流速转换,该线性因子可以通过珠子在低流速下的布朗运动或使用 DNA 过度拉伸过渡进行校准。此外,基于理论模型计算了由于珠上的剪切流引起的流体动力和扭矩以及 DNA 上的等效拉伸力,同时还考虑了 DNA 上的流体动力阻力。计算出的力-延伸曲线与实测曲线非常吻合。这些结果为基于流动池的单分子拉伸实验中的力提供了重要的见解,并为将流动池建立为一种简单、低成本、灵活且精确的工具,用于在广泛的力范围内进行单分子力测量提供了基础范围。
更新日期:2021-09-15
中文翻译:

量化基于流动池的单分子拉伸实验中的力
基于流动池的单分子操作技术在 DNA 力学和蛋白质-DNA 相互作用的研究中有许多应用。然而,这些实验中的力尚未完全表征,通常仅限于中等力制度 (<25 pN)。在这项工作中,使用“系留珠”测定法,基于“珠-弹簧链”模型定量评估了 DNA 的流体动力学阻力。因此,源自珠子布朗运动的力既包含来自 DNA 的等效流体动力学阻力的贡献,也包含来自系留珠子的拉力。接下来,使用基于流动池的 DNA 拉动实验,流量和珠子 DNA 系统上的总流体动力之间的线性关系已被证明在很宽的力范围 (0–110 pN) 内是有效的。最后,力可以通过线性因子直接从流速转换,该线性因子可以通过珠子在低流速下的布朗运动或使用 DNA 过度拉伸过渡进行校准。此外,基于理论模型计算了由于珠上的剪切流引起的流体动力和扭矩以及 DNA 上的等效拉伸力,同时还考虑了 DNA 上的流体动力阻力。计算出的力-延伸曲线与实测曲线非常吻合。这些结果为基于流动池的单分子拉伸实验中的力提供了重要的见解,并为将流动池建立为一种简单、低成本、灵活且精确的工具,用于在广泛的力范围内进行单分子力测量提供了基础范围。











































 京公网安备 11010802027423号
京公网安备 11010802027423号