当前位置:
X-MOL 学术
›
ACS Synth. Biol.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
A Platform for Deep Sequence–Activity Mapping and Engineering Antimicrobial Peptides
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2021-09-10 , DOI: 10.1021/acssynbio.1c00314 Matthew P DeJong 1 , Seth C Ritter 1 , Katharina A Fransen 1 , Daniel T Tresnak 1 , Alexander W Golinski 1 , Benjamin J Hackel 1
ACS Synthetic Biology ( IF 3.7 ) Pub Date : 2021-09-10 , DOI: 10.1021/acssynbio.1c00314 Matthew P DeJong 1 , Seth C Ritter 1 , Katharina A Fransen 1 , Daniel T Tresnak 1 , Alexander W Golinski 1 , Benjamin J Hackel 1
Affiliation
|
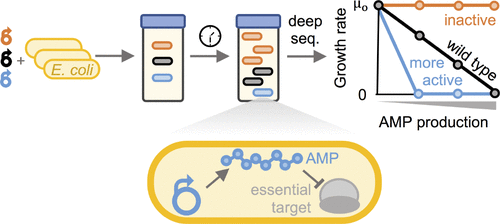
Developing potent antimicrobials, and platforms for their study and engineering, is critical as antibiotic resistance grows. A high-throughput method to quantify antimicrobial peptide and protein (AMP) activity across a broad continuum would be powerful to elucidate sequence–activity landscapes and identify potent mutants. Yet the complexity of antimicrobial activity has largely constrained the scope and mechanistic bandwidth of AMP variant analysis. We developed a platform to efficiently perform sequence–activity mapping of AMPs via depletion (SAMP-Dep): a bacterial host culture is transformed with an AMP mutant library, induced to intracellularly express AMPs, grown under selective pressure, and deep sequenced to quantify mutant depletion. The slope of mutant growth rate versus induction level indicates potency. Using SAMP-Dep, we mapped the sequence–activity landscape of 170 000 mutants of oncocin, a proline-rich AMP, for intracellular activity against Escherichia coli. Clonal validation supported the platform’s sensitivity and accuracy. The mapped landscape revealed an extended oncocin pharmacophore contrary to earlier structural studies, clarified the C-terminus role in internalization, identified functional epistasis, and guided focused, successful synthetic peptide library design, yielding a mutant with 2-fold enhancement in both intracellular and extracellular activity. The efficiency of SAMP-Dep poises the platform to transform AMP engineering, characterization, and discovery.
中文翻译:

深度序列-活性图谱和工程抗菌肽的平台
随着抗生素耐药性的增加,开发有效的抗菌剂及其研究和工程平台至关重要。一种在广泛的连续统一体中量化抗菌肽和蛋白质 (AMP) 活性的高通量方法将有力地阐明序列-活性景观和识别有效的突变体。然而,抗菌活性的复杂性在很大程度上限制了 AMP 变异分析的范围和机制带宽。我们开发了一个平台,通过消耗 (SAMP-Dep):细菌宿主培养物用 AMP 突变文库转化,诱导细胞内表达 AMP,在选择性压力下生长,并深度测序以量化突变消耗。突变体生长速率与诱导水平的斜率表明效力。使用 SAMP-Dep,我们绘制了 170 000 个 oncocin 突变体(一种富含脯氨酸的 AMP)的序列-活性图谱,用于对抗大肠杆菌的细胞内活性. 克隆验证支持平台的灵敏度和准确性。映射的景观揭示了与早期结构研究相反的扩展的癌素药效团,阐明了 C 末端在内化中的作用,确定了功能上位性,并指导了集中的、成功的合成肽库设计,产生了在细胞内和细胞外均增强 2 倍的突变体活动。SAMP-Dep 的效率使平台能够转变 AMP 工程、表征和发现。
更新日期:2021-10-15
中文翻译:

深度序列-活性图谱和工程抗菌肽的平台
随着抗生素耐药性的增加,开发有效的抗菌剂及其研究和工程平台至关重要。一种在广泛的连续统一体中量化抗菌肽和蛋白质 (AMP) 活性的高通量方法将有力地阐明序列-活性景观和识别有效的突变体。然而,抗菌活性的复杂性在很大程度上限制了 AMP 变异分析的范围和机制带宽。我们开发了一个平台,通过消耗 (SAMP-Dep):细菌宿主培养物用 AMP 突变文库转化,诱导细胞内表达 AMP,在选择性压力下生长,并深度测序以量化突变消耗。突变体生长速率与诱导水平的斜率表明效力。使用 SAMP-Dep,我们绘制了 170 000 个 oncocin 突变体(一种富含脯氨酸的 AMP)的序列-活性图谱,用于对抗大肠杆菌的细胞内活性. 克隆验证支持平台的灵敏度和准确性。映射的景观揭示了与早期结构研究相反的扩展的癌素药效团,阐明了 C 末端在内化中的作用,确定了功能上位性,并指导了集中的、成功的合成肽库设计,产生了在细胞内和细胞外均增强 2 倍的突变体活动。SAMP-Dep 的效率使平台能够转变 AMP 工程、表征和发现。











































 京公网安备 11010802027423号
京公网安备 11010802027423号