Bioorganic & Medicinal Chemistry ( IF 3.5 ) Pub Date : 2021-09-04 , DOI: 10.1016/j.bmc.2021.116393 Yash Gupta 1 , Sumit Kumar 2 , Samantha E Zak 3 , Krysten A Jones 4 , Charu Upadhyay 2 , Neha Sharma 5 , Saara-Anne Azizi 4 , Rahul S Kathayat 4 , Poonam 2 , Andrew S Herbert 6 , Ravi Durvasula 1 , Bryan C Dickinson 4 , John M Dye 3 , Brijesh Rathi 5 , Prakasha Kempaiah 1
|
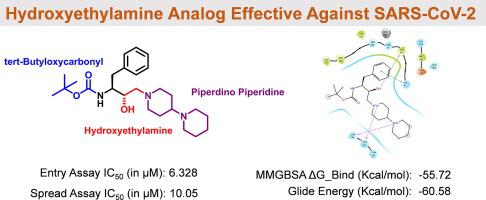
The continued toll of COVID-19 has halted the smooth functioning of civilization on a global scale. With a limited understanding of all the essential components of viral machinery and the lack of structural information of this new virus, initial drug discovery efforts had limited success. The availability of high-resolution crystal structures of functionally essential SARS-CoV-2 proteins, including 3CLpro, supports the development of target-specific therapeutics. 3CLpro, the main protease responsible for the processing of viral polypeptide, plays a vital role in SARS-CoV-2 viral replication and translation and is an important target in other coronaviruses. Additionally, 3CLpro is the target of repurposed drugs, such as lopinavir and ritonavir. In this study, target proteins were retrieved from the protein data bank (PDB IDs: 6 M03, 6LU7, 2GZ7, 6 W63, 6SQS, 6YB7, and 6YVF) representing different open states of the main protease to accommodate macromolecular substrate. A hydroxyethylamine (HEA) library was constructed from harvested chemical structures from all the series being used in our laboratories for screening against malaria and Leishmania parasites. The database consisted of ∼1000 structure entries, of which 70% were new to ChemSpider at the time of screening. This in-house library was subjected to high throughput virtual screening (HTVS), followed by standard precision (SP) and then extra precision (XP) docking (Schrodinger LLC 2021). The ligand strain and complex energy of top hits were calculated by Molecular Mechanics Generalized Born Surface Area (MM/GBSA) method. Promising hit compounds (n = 40) specifically binding to 3CLpro with high energy and average MM/GBSA scores were then subjected to (100-ns) MD simulations. Using this sequential selection followed by an in-silico validation approach, we found a promising HEA-based compound (N,N'-((3S,3′S)-piperazine-1,4-diylbis(3-hydroxy-1-phenylbutane-4,2-diyl))bis(2-(5-methyl-1,3-dioxoisoindolin-2-yl)-3-phenylpropanamide)), which showed high in vitro antiviral activity against SARS-CoV-2. Further to reduce the size of the otherwise larger ligand, a pharmacophore-based predicted library of ∼42 derivatives was constructed, which were added to the previous compound library and rescreened virtually. Out of several hits from the predicted library, two compounds were synthesized, tested against SARS-CoV-2 culture, and found to have markedly improved antiviral activity.
中文翻译:

羟乙胺类似物的抗病毒评价:SARS-CoV-2 主要蛋白酶 (3CLpro) 的抑制剂,一种虚拟筛选和模拟方法
COVID-19 的持续造成的损失已经停止了全球范围内文明的平稳运转。由于对病毒机制的所有基本组成部分的了解有限,并且缺乏这种新病毒的结构信息,最初的药物发现工作取得了有限的成功。功能必需的 SARS-CoV-2 蛋白(包括 3CLpro)的高分辨率晶体结构的可用性支持靶向特异性疗法的开发。3CLpro 是负责病毒多肽加工的主要蛋白酶,在 SARS-CoV-2 病毒复制和翻译中起着至关重要的作用,是其他冠状病毒的重要靶标。此外,3CLpro 是再利用药物的目标,例如洛匹那韦和利托那韦。在这项研究中,目标蛋白是从蛋白质数据库(PDB IDs:6 M03、6LU7、2GZ7、6 W63、6SQS、6YB7 和 6YVF) 代表主要蛋白酶的不同开放状态以适应大分子底物。羟乙胺 (HEA) 文库是从我们实验室用于筛查疟疾和利什曼原虫寄生虫的所有系列中收集的化学结构构建而成的。该数据库包含约 1000 个结构条目,其中 70% 在筛选时对 ChemSpider 是新的。这个 其中 70% 在筛选时是 ChemSpider 的新手。这个 其中 70% 在筛选时是 ChemSpider 的新手。这个内部图书馆进行高通量虚拟筛选 (HTVS),然后进行标准精度 (SP) 和超精度 (XP) 对接 (Schrodinger LLC 2021)。通过分子力学广义出生表面积(MM / GBSA)方法计算顶部命中的配体应变和复合能量。然后对具有高能量和平均 MM/GBSA 分数的 3CLpro 特异性结合的有希望的命中化合物 (n = 40) 进行 (100-ns) MD 模拟。使用这种顺序选择和计算机验证方法,我们发现了一种很有前途的基于 HEA 的化合物 (N,N'-((3S,3′S)-piperazine-1,4-diylbis(3-hydroxy-1- phenylbutane-4,2-diyl))bis(2-(5-methyl-1,3-dioxoisoindolin-2-yl)-3-phenylpropanamide)),在体外表现出高对 SARS-CoV-2 的抗病毒活性。为了进一步减小原本较大的配体的大小,构建了一个基于药效团的约 42 种衍生物的预测库,将其添加到先前的化合物库中并进行虚拟重新筛选。在预测库的几个命中中,合成了两种化合物,对 SARS-CoV-2 培养物进行了测试,发现其抗病毒活性显着提高。



























 京公网安备 11010802027423号
京公网安备 11010802027423号