Protein & Peptide Letters ( IF 1.0 ) Pub Date : 2021-10-31 , DOI: 10.2174/0929866528666210903162137 Jaspreet Kaur Boparai 1 , Nancy 1 , Pushpender Kumar Sharma 1
|
Aim: This study was designed to screen and identify an antimicrobial peptide from rhizosphere soil. The study was further focused towards overexpression, purification and characterization of this antimicrobial peptide, and to functionally validate its efficiency and efficacy as an antimicrobial agent. Yet, the study was further aimed at corroborating structural and functional studies using biophysical tools.
Background: Antimicrobial resistance is emerging as one of the top 10 global health crisis, it is multifaceted and the second largest cause of mortality. According to the World Health Organization (WHO), around the world, an estimated 700,000 people die each year from infection caused by antibiotic-resistant microbes. Antimicrobial peptides offer the best alternative to combat and overcome this crisis. In this manuscript, we report cloning, expression, purification and characterization of an antimicrobial peptide discovered from rhizosphere soil.
Objective: Objectives of this study include construction, screening and identification of antimicrobial peptide from metagenome followed by its expression, purification and functional and biophysical investigation. Yet another objective of the study was to determine antimicrobial efficacy and efficiency as an antimicrobial peptide against MRSA strains.
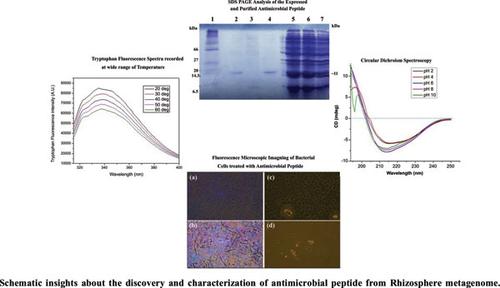
Methods: In this study, we used an array of molecular biology tools that include genetic engineering, PCR amplification, construction of an expression construct and NI-NTA based purification of the recombinant peptide. We have also carried out antimicrobial activity assay to determine MIC (minimum inhibitory concentration) and IC50 values of antimicrobial peptide. To establish the structural and functional relationship, circular dichroism, and both extrinsic and intrinsic fluorescence spectroscopy studies were carried out.
Results: Screening of metagenomic library resulted in the identification of gene (~500bp) harbouring an open reading frame (ORF) consisting of 282 bp. Open reading frame identified in gene encodes an antimicrobial peptide which had shared ~95% sequence similarity with the antimicrobial peptide of Bacillus origin. Purification of recombinant protein using Ni-NTA column chromatography demonstrated a purified protein band of ~11 kDa on 14% SDS-PAGE, which is well corroborated to theoretical deduced molecular weight of peptide from its amino acids sequence. Interestingly, the peptide exhibited antimicrobial activity in a broad range of pH and temperature. MIC determined against gram positive Bacillus sp. was found to be 0.015mg/ml, whereas, in the case of gram negative E. coli, it was calculated to be 0.062mg/ml. The peptide exhibited IC50 values corresponding to ~0.25mg/ml against Bacillus and ~0.5 mg/ml against E. coli. Antimicrobial susceptibility assay performed against methicillin resistant Staphylococcus aureus strain ATCC 3412 and standard strain of Staphylococcus aureus ATCC 9144 revealed its strong inhibitory activity against MRSA, whereby we observed a ~16mm clearance zone at higher peptide concentrations ~2mg/ml (~181.8μM). Biophysical investigation carried out using Trp fluorescence, ANS fluorescence and circular dichroism spectroscopy further revealed conformational stability in its secondary and tertiary structure at a wide range of temperature and pH.
Conclusion: Altogether, the peptide discovered from rhizosphere metagenome holds potential in inhibiting the growth of both gram positive and gram negative bacteria, and was equally effective in inhibiting the multidrug resistant pathogenic strains (MRSA).
中文翻译:

根际土壤抗菌肽的分子克隆、功能和生物物理表征
目的:本研究旨在筛选和鉴定根际土壤中的抗菌肽。该研究进一步侧重于这种抗菌肽的过表达、纯化和表征,并在功能上验证其作为抗菌剂的效率和功效。然而,该研究进一步旨在使用生物物理工具证实结构和功能研究。
背景:抗菌素耐药性正在成为全球十大健康危机之一,它是多方面的,也是导致死亡的第二大原因。根据世界卫生组织 (WHO) 的数据,全世界每年估计有 700,000 人死于由耐抗生素微生物引起的感染。抗菌肽提供了对抗和克服这一危机的最佳选择。在这份手稿中,我们报告了从根际土壤中发现的一种抗菌肽的克隆、表达、纯化和表征。
目的:本研究的目的包括从宏基因组构建、筛选和鉴定抗菌肽,然后对其表达、纯化以及功能和生物物理研究。该研究的另一个目的是确定作为抗 MRSA 菌株的抗微生物肽的抗微生物功效和效率。
方法:在这项研究中,我们使用了一系列分子生物学工具,包括基因工程、PCR 扩增、表达构建体的构建和基于 NI-NTA 的重组肽纯化。我们还进行了抗菌活性测定以确定抗菌肽的 MIC(最小抑制浓度)和 IC50 值。为了建立结构和功能关系,进行了圆二色性以及外在和内在荧光光谱研究。
结果:对宏基因组文库的筛选导致鉴定出具有由 282 bp 组成的开放阅读框 (ORF) 的基因 (~500bp)。在基因中鉴定的开放阅读框编码一种抗菌肽,该肽与芽孢杆菌来源的抗菌肽具有约 95% 的序列相似性。使用 Ni-NTA 柱层析纯化重组蛋白在 14% SDS-PAGE 上显示了约 11 kDa 的纯化蛋白条带,这很好地证实了从其氨基酸序列推导出的肽的理论分子量。有趣的是,该肽在广泛的 pH 值和温度范围内表现出抗菌活性。针对革兰氏阳性芽孢杆菌测定的 MIC。发现为 0.015mg/ml,而在革兰氏阴性大肠杆菌的情况下,计算为 0.062mg/ml。该肽对芽孢杆菌的 IC50 值约为 0.25mg/ml,对大肠杆菌的 IC50 值约为 0.5mg/ml。对耐甲氧西林金黄色葡萄球菌菌株 ATCC 3412 和标准金黄色葡萄球菌菌株 ATCC 9144 进行的抗菌药敏试验揭示了其对 MRSA 的强抑制活性,由此我们观察到在较高的肽浓度 ~2mg/ml (~181.8μM) 时有 ~16mm 的清除区。使用 Trp 荧光、ANS 荧光和圆二色光谱进行的生物物理研究进一步揭示了其二级和三级结构在很宽的温度和 pH 范围内的构象稳定性。对耐甲氧西林金黄色葡萄球菌菌株 ATCC 3412 和标准金黄色葡萄球菌菌株 ATCC 9144 进行的抗菌药敏试验揭示了其对 MRSA 的强抑制活性,由此我们观察到在较高的肽浓度 ~2mg/ml (~181.8μM) 时有 ~16mm 的清除区。使用 Trp 荧光、ANS 荧光和圆二色光谱进行的生物物理研究进一步揭示了其二级和三级结构在很宽的温度和 pH 范围内的构象稳定性。对耐甲氧西林金黄色葡萄球菌菌株 ATCC 3412 和标准金黄色葡萄球菌菌株 ATCC 9144 进行的抗菌药敏试验揭示了其对 MRSA 的强抑制活性,由此我们观察到在较高的肽浓度 ~2mg/ml (~181.8μM) 时有 ~16mm 的清除区。使用 Trp 荧光、ANS 荧光和圆二色光谱进行的生物物理研究进一步揭示了其二级和三级结构在很宽的温度和 pH 范围内的构象稳定性。
结论:总而言之,从根际宏基因组中发现的肽具有抑制革兰氏阳性和革兰氏阴性细菌生长的潜力,并且在抑制多重耐药致病菌株(MRSA)方面同样有效。











































 京公网安备 11010802027423号
京公网安备 11010802027423号