Journal of Molecular Biology ( IF 4.7 ) Pub Date : 2021-09-03 , DOI: 10.1016/j.jmb.2021.167229 Bi Zhao 1 , Akila Katuwawala 1 , Christopher J Oldfield 2 , Gang Hu 3 , Zhonghua Wu 4 , Vladimir N Uversky 5 , Lukasz Kurgan 1
|
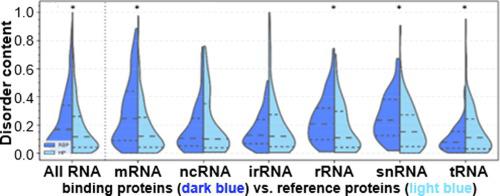
Although RNA-binding proteins (RBPs) are known to be enriched in intrinsic disorder, no previous analysis focused on RBPs interacting with specific RNA types. We fill this gap with a comprehensive analysis of the putative disorder in RBPs binding to six common RNA types: messenger RNA (mRNA), transfer RNA (tRNA), small nuclear RNA (snRNA), non-coding RNA (ncRNA), ribosomal RNA (rRNA), and internal ribosome RNA (irRNA). We also analyze the amount of putative intrinsic disorder in the RNA-binding domains (RBDs) and non-RNA-binding-domain regions (non-RBD regions). Consistent with previous studies, we show that in comparison with human proteome, RBPs are significantly enriched in disorder. However, closer examination finds significant enrichment in predicted disorder for the mRNA-, rRNA- and snRNA-binding proteins, while the proteins that interact with ncRNA and irRNA are not enriched in disorder, and the tRNA-binding proteins are significantly depleted in disorder. We show a consistent pattern of significant disorder enrichment in the non-RBD regions coupled with low levels of disorder in RBDs, which suggests that disorder is relatively rarely utilized in the RNA-binding regions. Our analysis of the non-RBD regions suggests that disorder harbors posttranslational modification sites and is involved in the putative interactions with DNA. Importantly, we utilize experimental data from DisProt and independent data from Pfam to validate the above observations that rely on the disorder predictions. This study provides new insights into the distribution of disorder across proteins that bind different RNA types and the functional role of disorder in the regions where it is enriched.
中文翻译:

人类 RNA 结合蛋白的内在障碍
尽管已知 RNA 结合蛋白 (RBP) 富含内在障碍,但之前的分析并未集中在 RBP 与特定 RNA 类型的相互作用上。我们通过全面分析 RBP 与六种常见 RNA 类型结合的假定障碍来填补这一空白:信使 RNA (mRNA)、转移 RNA (tRNA)、小核 RNA (snRNA)、非编码 RNA (ncRNA)、核糖体 RNA (rRNA) 和内部核糖体 RNA (irRNA)。我们还分析了 RNA 结合域 (RBD) 和非 RNA 结合域区域(非 RBD 区域)中假定的内在障碍的数量。与之前的研究一致,我们表明与人类蛋白质组相比,RBPs 在紊乱中显着丰富。然而,仔细检查发现 mRNA-、rRNA- 和 snRNA 结合蛋白的预测疾病显着富集,而与 ncRNA 和 irRNA 相互作用的蛋白质没有在无序状态下富集,而 tRNA 结合蛋白在无序状态下显着耗尽。我们在非 RBD 区域显示出一致的显着紊乱富集模式以及 RBD 中低水平的紊乱,这表明在 RNA 结合区域中相对很少利用紊乱。我们对非 RBD 区域的分析表明,这种紊乱具有翻译后修饰位点,并参与了与 DNA 的假定相互作用。重要的是,我们利用来自 DisProt 的实验数据和来自 Pfam 的独立数据来验证上述依赖于无序预测的观察结果。










































 京公网安备 11010802027423号
京公网安备 11010802027423号