当前位置:
X-MOL 学术
›
NMR Biomed.
›
论文详情
Our official English website, www.x-mol.net, welcomes your
feedback! (Note: you will need to create a separate account there.)
Automated segmentation of biventricular contours in tissue phase mapping using deep learning
NMR in Biomedicine ( IF 2.7 ) Pub Date : 2021-09-02 , DOI: 10.1002/nbm.4606 Daming Shen 1, 2 , Ashitha Pathrose 1 , Roberto Sarnari 1 , Allison Blake 1 , Haben Berhane 1, 2 , Justin J Baraboo 1, 2 , James C Carr 1 , Michael Markl 1, 2 , Daniel Kim 1, 2
NMR in Biomedicine ( IF 2.7 ) Pub Date : 2021-09-02 , DOI: 10.1002/nbm.4606 Daming Shen 1, 2 , Ashitha Pathrose 1 , Roberto Sarnari 1 , Allison Blake 1 , Haben Berhane 1, 2 , Justin J Baraboo 1, 2 , James C Carr 1 , Michael Markl 1, 2 , Daniel Kim 1, 2
Affiliation
|
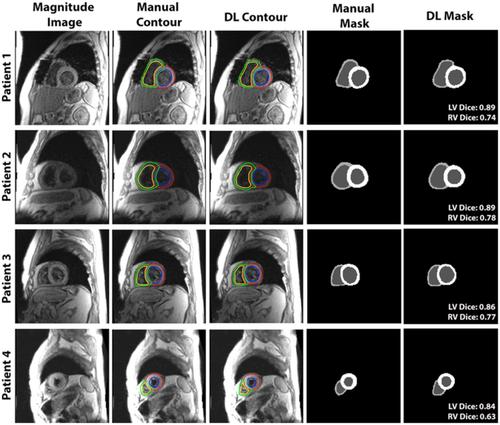
Tissue phase mapping (TPM) is an MRI technique for quantification of regional biventricular myocardial velocities. Despite its potential, clinical use is limited due to the requisite labor-intensive manual segmentation of cardiac contours for all time frames. The purpose of this study was to develop a deep learning (DL) network for automated segmentation of TPM images, without significant loss in segmentation and myocardial velocity quantification accuracy compared with manual segmentation. We implemented a multi-channel 3D (three dimensional; 2D + time) dense U-Net that trained on magnitude and phase images and combined cross-entropy, Dice, and Hausdorff distance loss terms to improve the segmentation accuracy and suppress unnatural boundaries. The dense U-Net was trained and tested with 150 multi-slice, multi-phase TPM scans (114 scans for training, 36 for testing) from 99 heart transplant patients (44 females, 1-4 scans/patient), where the magnitude and velocity-encoded (Vx, Vy, Vz) images were used as input and the corresponding manual segmentation masks were used as reference. The accuracy of DL segmentation was evaluated using quantitative metrics (Dice scores, Hausdorff distance) and linear regression and Bland-Altman analyses on the resulting peak radial and longitudinal velocities (Vr and Vz). The mean segmentation time was about 2 h per patient for manual and 1.9 ± 0.3 s for DL. Our network produced good accuracy (median Dice = 0.85 for left ventricle (LV), 0.64 for right ventricle (RV), Hausdorff distance = 3.17 pixels) compared with manual segmentation. Peak Vr and Vz measured from manual and DL segmentations were strongly correlated (R ≥ 0.88) and in good agreement with manual analysis (mean difference and limits of agreement for Vz and Vr were −0.05 ± 0.98 cm/s and −0.06 ± 1.18 cm/s for LV, and −0.21 ± 2.33 cm/s and 0.46 ± 4.00 cm/s for RV, respectively). The proposed multi-channel 3D dense U-Net was capable of reducing the segmentation time by 3,600-fold, without significant loss in accuracy in tissue velocity measurements.
中文翻译:

使用深度学习在组织相位映射中自动分割双心室轮廓
组织相位映射 (TPM) 是一种用于量化局部双心室心肌速度的 MRI 技术。尽管具有潜力,但由于在所有时间范围内对心脏轮廓进行必要的劳动密集型手动分割,临床使用受到限制。本研究的目的是开发一种深度学习 (DL) 网络,用于 TPM 图像的自动分割,与手动分割相比,分割和心肌速度量化精度没有显着损失。我们实现了一个多通道 3D(三维;2D + 时间)密集 U-Net,它在幅度和相位图像上进行训练,并结合交叉熵、Dice 和 Hausdorff 距离损失项来提高分割精度并抑制不自然的边界。密集的 U-Net 使用 150 个多切片进行训练和测试,V x , V y , V z ) 图像用作输入,相应的手动分割掩码用作参考。使用定量指标(Dice 分数、Hausdorff 距离)和线性回归以及 Bland-Altman 分析对所得峰值径向和纵向速度( V r和V z )评估 DL 分割的准确性。手动的平均分割时间约为每名患者 2 小时,DL 为 1.9 ± 0.3 秒。与手动分割相比,我们的网络产生了良好的准确性(左心室 (LV) 的中位数 Dice = 0.85,右心室 (RV) 的中位数 Dice = 0.64,Hausdorff 距离 = 3.17 像素)。峰值V r和从手动和 DL 分割测量的V z高度相关 ( R ≥ 0.88) 并且与手动分析非常一致(V z和V r的平均差异和一致性限制分别为 -0.05 ± 0.98 cm/s 和 -0.06 ± 1.18 cm /s 为 LV,分别为 -0.21 ± 2.33 cm/s 和 RV 为 0.46 ± 4.00 cm/s)。所提出的多通道 3D 密集 U-Net 能够将分割时间减少 3,600 倍,而不会显着降低组织速度测量的准确性。
更新日期:2021-09-02
中文翻译:

使用深度学习在组织相位映射中自动分割双心室轮廓
组织相位映射 (TPM) 是一种用于量化局部双心室心肌速度的 MRI 技术。尽管具有潜力,但由于在所有时间范围内对心脏轮廓进行必要的劳动密集型手动分割,临床使用受到限制。本研究的目的是开发一种深度学习 (DL) 网络,用于 TPM 图像的自动分割,与手动分割相比,分割和心肌速度量化精度没有显着损失。我们实现了一个多通道 3D(三维;2D + 时间)密集 U-Net,它在幅度和相位图像上进行训练,并结合交叉熵、Dice 和 Hausdorff 距离损失项来提高分割精度并抑制不自然的边界。密集的 U-Net 使用 150 个多切片进行训练和测试,V x , V y , V z ) 图像用作输入,相应的手动分割掩码用作参考。使用定量指标(Dice 分数、Hausdorff 距离)和线性回归以及 Bland-Altman 分析对所得峰值径向和纵向速度( V r和V z )评估 DL 分割的准确性。手动的平均分割时间约为每名患者 2 小时,DL 为 1.9 ± 0.3 秒。与手动分割相比,我们的网络产生了良好的准确性(左心室 (LV) 的中位数 Dice = 0.85,右心室 (RV) 的中位数 Dice = 0.64,Hausdorff 距离 = 3.17 像素)。峰值V r和从手动和 DL 分割测量的V z高度相关 ( R ≥ 0.88) 并且与手动分析非常一致(V z和V r的平均差异和一致性限制分别为 -0.05 ± 0.98 cm/s 和 -0.06 ± 1.18 cm /s 为 LV,分别为 -0.21 ± 2.33 cm/s 和 RV 为 0.46 ± 4.00 cm/s)。所提出的多通道 3D 密集 U-Net 能够将分割时间减少 3,600 倍,而不会显着降低组织速度测量的准确性。









































 京公网安备 11010802027423号
京公网安备 11010802027423号