Microbial Pathogenesis ( IF 3.8 ) Pub Date : 2021-09-02 , DOI: 10.1016/j.micpath.2021.105171 Jyotirmayee Dey 1 , Soumya Ranjan Mahapatra 1 , Pratima Singh 2 , Swadheena Patro 3 , Gajraj Singh Kushwaha 4 , Namrata Misra 5 , Mrutyunjay Suar 5
|
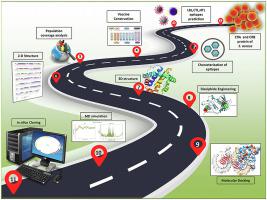
Staphylococcus aureus infection is emerging as a global threat because of the highly debilitating nature of the associated disease's unprecedented magnitude of its spread and growing global resistance to antimicrobial medicines. Recently WHO has categorized these bacteria under the high global priority pathogen list and is one of the six nosocomial pathogens termed as ESKAPE pathogens which have emerged as a serious threat to public health worldwide. The development of a specific vaccine can stimulate an optimal antibody response, thus providing immunity against it. Therefore, in the present study efforts have been made to identify potential vaccine candidates from the Clumping factor surface proteins (ClfA and ClfB) of S. aureus. Employing the immunoinformatics approach, fourteen antigenic peptides including T-cell, B-cell epitopes were identified which were non-toxic, non-allergenic, high antigenicity, strong binding efficiency with commonly occurring MHC alleles. Consequently, a multi-epitope vaccine chimera was designed by connecting these epitopes with suitable linkers an adjuvant to enhance immunogenicity. Further, homology modeling and molecular docking were performed to construct the three-dimensional structure of the vaccine and study the interaction between the modeled structure and immune receptor (TLR-2) present on lymphocyte cells. Consequently, molecular dynamics simulation for 100 ns period confirmed the stability of the interaction and reliability of the structure for further analysis. Finally, codon optimization and in silico cloning were employed to ensure the successful expression of the vaccine candidate. As the targeted protein is highly antigenic and conserved, hence the designed novel vaccine construct holds potential against emerging multi-drug-resistant organisms.
中文翻译:

从金黄色葡萄球菌的凝集因子蛋白预测的基于 B 和 T 细胞表位的肽作为疫苗靶标
金黄色葡萄球菌感染正在成为一种全球性威胁,因为相关疾病具有前所未有的传播规模以及全球对抗菌药物的耐药性日益增强的高度衰弱性。最近,世卫组织将这些细菌归类为全球高度优先病原体清单,是被称为 ESKAPE 病原体的六种医院病原体之一,已成为对全球公共卫生的严重威胁。特定疫苗的开发可以刺激最佳的抗体反应,从而提供针对它的免疫力。因此,在本研究中,已努力从金黄色葡萄球菌的聚集因子表面蛋白(ClfA 和 ClfB)中鉴定潜在的候选疫苗。. 采用免疫信息学方法,鉴定出14种抗原肽,包括T细胞、B细胞表位,这些抗原肽无毒、无过敏性、高抗原性、与常见的MHC等位基因结合效率高。因此,通过将这些表位与合适的接头和佐剂连接来设计多表位疫苗嵌合体以增强免疫原性。此外,通过同源建模和分子对接,构建了疫苗的三维结构,并研究了建模结构与淋巴细胞上存在的免疫受体(TLR-2)之间的相互作用。因此,100 ns 周期的分子动力学模拟证实了相互作用的稳定性和结构的可靠性,以供进一步分析。最后,密码子优化和计算机克隆被用来确保候选疫苗的成功表达。由于靶向蛋白具有高度抗原性和保守性,因此设计的新型疫苗构建体具有对抗新兴多药耐药生物的潜力。



























 京公网安备 11010802027423号
京公网安备 11010802027423号